 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Ciclev10027889m | ||||||||
| Common Name | CICLE_v10027890mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 736aa MW: 78877.7 Da PI: 6.2995 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 50.6 | 3.3e-16 | 469 | 515 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn ErrRRdriN+++ L+el+P++ K++Ka++L +A+eY+k Lq
Ciclev10027889m 469 VHNLSERRRRDRINEKMRALQELIPNC-----NKVDKASMLDEAIEYLKTLQ 515
6*************************8.....6******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| CDD | cd00083 | 2.62E-17 | 461 | 519 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 3.6E-20 | 462 | 523 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 1.31E-20 | 462 | 527 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 18.349 | 465 | 514 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 1.1E-13 | 469 | 515 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 1.7E-17 | 471 | 520 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009704 | Biological Process | de-etiolation | ||||
| GO:0009740 | Biological Process | gibberellic acid mediated signaling pathway | ||||
| GO:0010017 | Biological Process | red or far-red light signaling pathway | ||||
| GO:0031539 | Biological Process | positive regulation of anthocyanin metabolic process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 736 aa Download sequence Send to blast |
MPLFELYQMT KEKLDSAKQK NPSSSTDLSF EPENDLVELV WENGQILKQG QSSRTKKSPT 60 SNSLPSYCLR SHTPKNRDKD LGKTGKFGTV DAMLNGIQMS DPSGEMGLNQ DDDMVPWLNY 120 AVDESLQQDY CSDFLHELSG VTGNELSSQN NFASFDKRSS SNQFDREFNS VFGNHGARLE 180 HGNVSKVSSV GGGEATRPRS STNSVYTSSS QQCQTSLPNL RSGVSDSTSN GIKNSTDNGI 240 FQGSNPTATG NLSGLKLHKQ DQRLSANNSG VMNFSHFARP VALVRANLQN IGSMPGARLS 300 RIERMGSKDK SSVASRSPVE SPLIDSSGCL RKESSCQPAA VTSKVDVKPQ EAKSSEQPVS 360 EPPEAACRED ALNNDKKPSQ VHAESATKGV VESEKTVEPV VAASSVCSGN SVERASDDLP 420 HNVKRKHCDL DDSECPSEEQ DVEEESVGVK KQVPGRVGTG SKRSRAAEVH NLSERRRRDR 480 INEKMRALQE LIPNCNKVDK ASMLDEAIEY LKTLQLQVQI MSMGAGFYMP PMMYPSGMQH 540 MHAAHMAHFP PMGIGMGMGI GYGMGMPDIN GGSSCYPMVQ VPAMHGAHFP GTSVSGPSAL 600 QGMAGSNFQL LGLPAQGHHM SMPHAPLNPL SGGPIMKSAM GLNPTGMVGS MDNTDAAKGS 660 TSKDLVQNVN SQVKQNTGAK GPVDQASGQC EATNESSAEP AAVQDNCQAS EVNGLVTYNS 720 SRKNENESSN PAFFE* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 473 | 478 | ERRRRD |
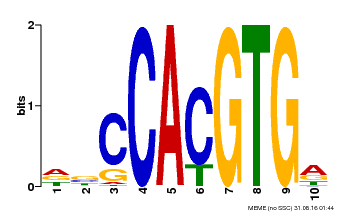
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00081 | ChIP-seq | Transfer from AT1G09530 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006423962.1 | 0.0 | transcription factor PIF3 | ||||
| Refseq | XP_006480339.1 | 0.0 | transcription factor PIF3 | ||||
| TrEMBL | V4SJJ6 | 0.0 | V4SJJ6_9ROSI; Uncharacterized protein | ||||
| STRING | XP_006480339.1 | 0.0 | (Citrus sinensis) | ||||
| STRING | XP_006423961.1 | 0.0 | (Citrus clementina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM7665 | 27 | 39 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09530.2 | 1e-54 | phytochrome interacting factor 3 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Ciclev10027889m |
| Entrez Gene | 18034179 |



