 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Ciclev10030185m | ||||||||
| Common Name | CICLE_v10030185mg | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Sapindales; Rutaceae; Aurantioideae; Citrus
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 497aa MW: 54724.1 Da PI: 6.0154 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 106.6 | 1.4e-33 | 265 | 319 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kpr+rWtpeLHe+Fveav++L G+ekAtPk++l+lm+v+gLt++hvkSHLQkYRl
Ciclev10030185m 265 KPRMRWTPELHECFVEAVNKLDGPEKATPKAVLKLMNVEGLTIYHVKSHLQKYRL 319
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 11.419 | 262 | 322 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 4.3E-18 | 263 | 319 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 7.7E-32 | 264 | 320 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 8.1E-26 | 265 | 320 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 4.1E-10 | 267 | 318 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 3.0E-21 | 354 | 400 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 497 aa Download sequence Send to blast |
MNHHSIISVT KNESNKGVSQ SCCSALSPIH NFQTEGQSLS TGEYPFPHPS PFIRKESLSS 60 PNRMQASTVV PKENGLISTL DSPISPGSHF QHSKGGFSRS SVFCTSLYLS SSASSETHRQ 120 IGNFPFLPHP RTFNQSVSAV DSTKSSLLFS EDMGNAYQEE HSESLMKGFL NFPEDASDGS 180 FPGVTCMGER LGLNEHLELQ FLSDELDIDI TDHGENPRLD EIDDAPKSSL EPPMGLSCNE 240 NYVSSAPPVD ALSSHTSPAS ATAHKPRMRW TPELHECFVE AVNKLDGPEK ATPKAVLKLM 300 NVEGLTIYHV KSHLQKYRLA KYMPEKKEEK KTCSSEEKKA TSSIESDGRK KGSIQFTEAL 360 RMQMEVQKQL HEQLEVQRAL QLRIEEHARY LEKIVAEQQK DGSATILPQA QSLSTITNGS 420 KDSEQQPSPP SFTVSAILSP EQPAESKTES SSTSLLSKHK ATDSRESKPD ACLKRIRLEN 480 KPEITSDEAV VENPVQ* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 1e-25 | 265 | 322 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 1e-25 | 265 | 322 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 1e-25 | 265 | 322 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 1e-25 | 265 | 322 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 1e-25 | 265 | 322 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 1e-25 | 265 | 322 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 1e-25 | 265 | 322 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 1e-25 | 265 | 322 | 2 | 59 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00354 | DAP | Transfer from AT3G13040 | Download |
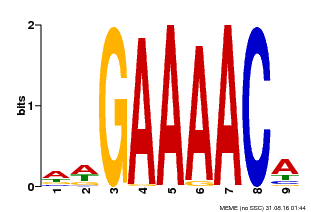
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | - | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_006425193.1 | 0.0 | myb family transcription factor PHL6 | ||||
| Refseq | XP_024034679.1 | 0.0 | myb family transcription factor PHL6 | ||||
| Swissprot | Q949U2 | 1e-135 | PHL6_ARATH; Myb family transcription factor PHL6 | ||||
| TrEMBL | V4RUQ0 | 0.0 | V4RUQ0_9ROSI; Uncharacterized protein | ||||
| STRING | XP_006425193.1 | 0.0 | (Citrus clementina) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6786 | 27 | 44 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G13040.2 | 1e-112 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Ciclev10030185m |
| Entrez Gene | 18034354 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



