 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cla000703 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Citrullus
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 415aa MW: 45440.5 Da PI: 4.7919 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 49.4 | 8e-16 | 176 | 222 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn E+rRR+riN+++ L+ l+P++ K +Ka++L +A+eY+k+Lq
Cla000703 176 VHNLSEKRRRSRINEKMKALQNLIPNS-----NKTDKASMLDEAIEYLKQLQ 222
6*************************7.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF47459 | 2.88E-20 | 169 | 232 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 3.5E-20 | 169 | 232 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.86 | 172 | 221 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 3.07E-17 | 175 | 226 | No hit | No description |
| Pfam | PF00010 | 2.7E-13 | 176 | 222 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 8.3E-18 | 178 | 227 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010114 | Biological Process | response to red light | ||||
| GO:0010154 | Biological Process | fruit development | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 415 aa Download sequence Send to blast |
MDHSIPNNCD GNACTSSTWT LPTAAVSSSS PPDDISLFLQ QILLRSSSSA PHSSLLSPSP 60 SIFSELTCNI RAFTPSTHNI PPPYGPSNAV PDEISAVDSS EQFANSSSSG VLHDPLRSCP 120 TPIPPNASST SVGASDHNEN DEFDCESEEG LEALVEEQPT KPNPRSSSKR SRAAEVHNLS 180 EKRRRSRINE KMKALQNLIP NSNKTDKASM LDEAIEYLKQ LQLQVQMLSM RNGLSLHPMN 240 LPGSLQYLQL SHMRMDFGEE NSSMSSDQER PNQIFLSLPD QKTASIHPFM SDIGRTNAET 300 PFELTPPIQA QLVPFYLSES SKSKEICSRD VLRGDHQVNM NVSQSETTPF MSHPYNISAP 360 PDLQGLKNCV SVEPSIIEGN RSGVLLNYTP EHSLVFPSPL NGIDTGRPTE TTELQ |
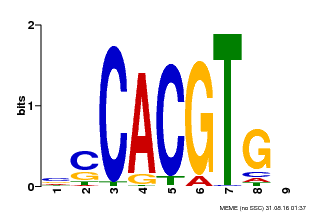
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00035 | PBM | Transfer from AT4G36930 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681875 | 1e-162 | LN681875.1 Cucumis melo genomic scaffold, anchoredscaffold00007. | |||
| GenBank | LN713262 | 1e-162 | LN713262.1 Cucumis melo genomic chromosome, chr_8. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004142005.2 | 0.0 | PREDICTED: transcription factor SPATULA | ||||
| TrEMBL | A0A0A0KLD3 | 0.0 | A0A0A0KLD3_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004157205.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2276 | 33 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G36930.1 | 2e-39 | bHLH family protein | ||||



