 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cla002428 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Citrullus
|
||||||||
| Family | TCP | ||||||||
| Protein Properties | Length: 432aa MW: 46917.3 Da PI: 6.9235 | ||||||||
| Description | TCP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCP | 121.2 | 1.2e-37 | 37 | 139 | 2 | 89 |
TCP 2 agkkdrhskihTkvggRdRRvRlsaecaarfFdLqdeLGfdkdsktieWLlqqakpaikeltgtssssasec...eaesssssasnsssg.......... 88
+g+kdrhsk++T++g+RdRRvRlsa++a++f+d+qd+LG+d++sk+++WL+++ak+ai+el+++++ ++s ++ ++++s + ++++
Cla002428 37 TGRKDRHSKVCTAKGPRDRRVRLSAHTAIQFYDVQDRLGYDRPSKAVDWLIKKAKAAIDELAELPAWHPSTGntsTQREEQQSLNDENENllsvqrevfn 136
79**************************************************************666663332223333333333333335555544444 PP
TCP 89 ..k 89
Cla002428 137 ttG 139
441 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF03634 | 7.6E-32 | 37 | 148 | IPR005333 | Transcription factor, TCP |
| PROSITE profile | PS51369 | 34.295 | 39 | 97 | IPR017887 | Transcription factor TCP subgroup |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009793 | Biological Process | embryo development ending in seed dormancy | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0030154 | Biological Process | cell differentiation | ||||
| GO:0045962 | Biological Process | positive regulation of development, heterochronic | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 432 aa Download sequence Send to blast |
MGESHRQTST SSRLGMRTGG GGGEIVEVQG GHIVRSTGRK DRHSKVCTAK GPRDRRVRLS 60 AHTAIQFYDV QDRLGYDRPS KAVDWLIKKA KAAIDELAEL PAWHPSTGNT STQREEQQSL 120 NDENENLLSV QREVFNTTGN RRATVLGSES RVSEFPLQNL QHTQMGEGPN SSTSSFLPPS 180 LDSDSIADTI KSFFPIGTAA AETTSSSIQF QNYPQDLLSR TSSQNQDLRL SLQSFQDPIA 240 IHRHHHAQHQ SQGHQNEHVL FSGTAPLSGF DVTTAGWSEH NSLNPAEISR FPRITCWNAS 300 GAETGSGGGS SGHGGGGIRS AGYVFNSPQL PTALPPSQLM QPLFGENQFF SQRGPLQSSN 360 TPSIRAWIDP SLTPTDHQHH QMPPSIHQSS YTALGFASGG FSGFHIPTRI QGEEEHDGIS 420 DKPSSASSDS RH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zkt_A | 1e-20 | 44 | 97 | 1 | 54 | Putative transcription factor PCF6 |
| 5zkt_B | 1e-20 | 44 | 97 | 1 | 54 | Putative transcription factor PCF6 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor playing a pivotal role in the control of morphogenesis of shoot organs by negatively regulating the expression of boundary-specific genes such as CUC genes, probably through the induction of miRNA (e.g. miR164) (PubMed:12931144, PubMed:17307931). Required during early steps of embryogenesis (PubMed:15634699). Participates in ovule develpment (PubMed:25378179). Activates LOX2 expression by binding to the 5'-GGACCA-3' motif found in its promoter (PubMed:18816164). {ECO:0000269|PubMed:12931144, ECO:0000269|PubMed:15634699, ECO:0000269|PubMed:17307931, ECO:0000269|PubMed:18816164, ECO:0000269|PubMed:25378179}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00009 | PBM | Transfer from 929286 | Download |
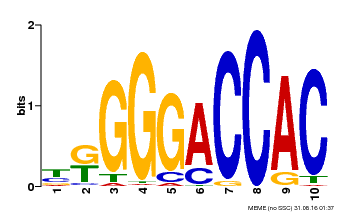
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by the miRNA miR-JAW/miR319 (PubMed:12931144, PubMed:18816164). {ECO:0000269|PubMed:12931144, ECO:0000269|PubMed:18816164}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681873 | 0.0 | LN681873.1 Cucumis melo genomic scaffold, anchoredscaffold00027. | |||
| GenBank | LN713261 | 0.0 | LN713261.1 Cucumis melo genomic chromosome, chr_7. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004137216.2 | 0.0 | PREDICTED: transcription factor TCP4, partial | ||||
| Swissprot | Q8LPR5 | 1e-104 | TCP4_ARATH; Transcription factor TCP4 | ||||
| TrEMBL | A0A0A0KXN1 | 0.0 | A0A0A0KXN1_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004137216.1 | 0.0 | (Cucumis sativus) | ||||
| STRING | XP_004172687.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1163 | 34 | 105 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G15030.3 | 4e-72 | TCP family protein | ||||



