 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cla004039 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Citrullus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 265aa MW: 30566.8 Da PI: 8.0741 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 44 | 5.1e-14 | 14 | 61 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+WT eEd +l + + G ++W+++++ g+ R++k+c++rw +yl
Cla004039 14 RGPWTVEEDHKLMNFILNNGIHCWRLVPKLAGLLRCGKSCRLRWINYL 61
89******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 56.1 | 8.3e-18 | 67 | 110 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
rg++T+ E+ ++++++ +lG++ W++Ia++++ gRt++++k++w++
Cla004039 67 RGAFTEMEENQIIHLHSLLGNR-WSKIAAHFP-GRTDNEIKNHWNT 110
89********************.*********.***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-21 | 5 | 64 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 14.011 | 9 | 61 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.02E-28 | 11 | 108 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.0E-8 | 13 | 63 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.9E-12 | 14 | 61 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.74E-6 | 16 | 61 | No hit | No description |
| PROSITE profile | PS51294 | 26.581 | 62 | 116 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.4E-26 | 65 | 117 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 9.9E-17 | 66 | 114 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.7E-16 | 67 | 110 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 6.65E-12 | 69 | 110 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 265 aa Download sequence Send to blast |
MGRQPCCDKV GLKRGPWTVE EDHKLMNFIL NNGIHCWRLV PKLAGLLRCG KSCRLRWINY 60 LRPDLKRGAF TEMEENQIIH LHSLLGNRWS KIAAHFPGRT DNEIKNHWNT RIKKRLKQLG 120 LDPLTHKKLT EKTDRLKEKL FIASNPFRKQ EAIMPNFDFE TKEINQVASH MISSDDGSSN 180 LVCKGIGYEM WTDQLENDSN VSFSLEYCSS MNESLSIIKQ NSIQFDSLDS LPSWDDGVPK 240 FNQLEKDDIY SLGNGGGYYV DTFSH |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 1e-28 | 12 | 116 | 5 | 108 | B-MYB |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that acts as positive regulator of abscisic acid (ABA) signaling in response to salt stress. Acts as negative regulator ABI1, ABI2 and PP2CA, which are protein phosphatases 2C acting as negative regulator of ABA signaling. Binds to the DNA specific sequence and core element 5'-ACGT-3' found in the promoters of ABI1 and PP2CA to negatively regulate their expression during ABA-dependent salt stress response. {ECO:0000269|PubMed:23660402}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00510 | DAP | Transfer from AT5G14340 | Download |
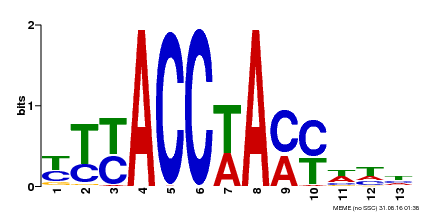
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by salt stress, drought stress and abscisic acid (ABA). Down-regulated by salicylic acid (SA) methyl jasmonate (JA). {ECO:0000269|PubMed:23660402}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681857 | 0.0 | LN681857.1 Cucumis melo genomic scaffold, anchoredscaffold00023. | |||
| GenBank | LN713260 | 0.0 | LN713260.1 Cucumis melo genomic chromosome, chr_6. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008450053.1 | 0.0 | PREDICTED: transcription repressor MYB6 | ||||
| Swissprot | Q9C7U7 | 5e-72 | MYB20_ARATH; Transcription factor MYB20 | ||||
| TrEMBL | A0A1S3BP24 | 0.0 | A0A1S3BP24_CUCME; transcription repressor MYB6 | ||||
| STRING | XP_008450053.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6625 | 31 | 50 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G14340.1 | 1e-82 | myb domain protein 40 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



