 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cla005462 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Citrullus
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 669aa MW: 75439.1 Da PI: 6.608 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 156.1 | 2.5e-48 | 60 | 195 | 1 | 134 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslq.sslk 99
gg+ +++++kE+E++k+RER+RRai+++i+aGLR++Gn++lp+raD+n+Vl+AL+reAGwvve+DGttyr+++ p ++++ ++ +s es+++ s k
Cla005462 60 GGKAKREREKEKERTKLRERHRRAITSRILAGLRQYGNFPLPARADMNDVLAALAREAGWVVEADGTTYRQSTPPS--QSQGAAFPVRSGESPISsGSFK 157
57899*****************************************************************999888..666777888888888887789* PP
DUF822 100 ssalaspvesysaspksssfpspssldsislasa...a 134
+++++ ++ + ++++ ++++sp+slds+ +++ +
Cla005462 158 GCSIKATLDCQPSVLRIDESLSPASLDSVVITERdakN 195
*****************************999876542 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 9.5E-49 | 61 | 205 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 3.86E-157 | 227 | 665 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 7.1E-169 | 230 | 661 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 5.3E-80 | 255 | 625 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-52 | 267 | 281 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-52 | 288 | 306 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-52 | 310 | 331 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-52 | 403 | 425 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-52 | 476 | 495 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-52 | 510 | 526 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-52 | 527 | 538 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-52 | 545 | 568 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.1E-52 | 583 | 605 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 669 aa Download sequence Send to blast |
MSGSLNDDSF HQDLQPQANH ASDYLSHQLQ PPPPPPRRPR GFAATAAAMG PTTTTAAATG 60 GKAKREREKE KERTKLRERH RRAITSRILA GLRQYGNFPL PARADMNDVL AALAREAGWV 120 VEADGTTYRQ STPPSQSQGA AFPVRSGESP ISSGSFKGCS IKATLDCQPS VLRIDESLSP 180 ASLDSVVITE RDAKNEKYTA LSPLNAAHCL EDQLIQDIRS RENESQFRGT PYVPVYVILA 240 TGFINNFCQL IDPDGVRQEL SHLQSLNVDG VIVDCWWGIV EAWNPQKYVW SGYRDLFNII 300 REFKLKVQVV MAFHAFGGTE SGDAFIKLPQ WVLEIGKENP DIFFTDREGR RNKDCLSWGI 360 DKERVLRGRT GIEVYFDFMR SFHTEFNDLF AEGLVSAIEV GLGAFGELKY PSFSERMGWR 420 YPGIGEFQCY DKYLQQSLRK AAGLRGHSFW ARGPDNAGQY NSRPHESGFF CERGDYDSYY 480 GRFFLQWYAQ TLIYHVDNVL SLASLVFEET KFIVKIPAVY WWYKTSSHAA ELTAGFYNPS 540 NQDGYSPVFD VLKKHCVIVK LVCCGMPVAG QEVDDTSADP ESLSWQILNS AWDRGLTVAG 600 ENSLSCYDRD GYMRIIDMAK PRSDPDRHHF SFFAYRQPSA LIQGAVCFPE LDYFIKCMHG 660 KYLICSSLF |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-122 | 232 | 667 | 11 | 450 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
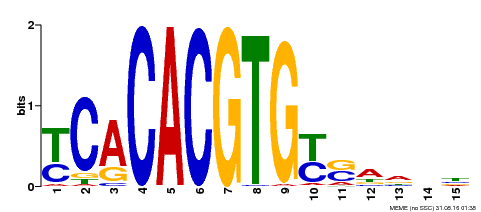
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681873 | 0.0 | LN681873.1 Cucumis melo genomic scaffold, anchoredscaffold00027. | |||
| GenBank | LN713261 | 0.0 | LN713261.1 Cucumis melo genomic chromosome, chr_7. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_022931287.1 | 0.0 | beta-amylase 8-like | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A1S3BTA5 | 0.0 | A0A1S3BTA5_CUCME; Beta-amylase | ||||
| STRING | XP_008451866.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11902 | 30 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.2 | 0.0 | beta-amylase 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



