 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cla006955 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Citrullus
|
||||||||
| Family | LBD | ||||||||
| Protein Properties | Length: 240aa MW: 24652 Da PI: 7.5021 | ||||||||
| Description | LBD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF260 | 129.6 | 1.3e-40 | 29 | 129 | 1 | 100 |
DUF260 1 aCaaCkvlrrkCakdCvlapyfpaeq.pkkfanvhklFGasnvlkllkalpeeeredamsslvyeAearardPvyGavgvilklqqqleqlkaelallke 99
+C aCk+lrrkC+++C++apyf +eq +++fa+vhk+FGasnv+kll ++p ++r da+ +++yeA+ar+rdPvyG+v++i++lqqq+ +l+ael +l++
Cla006955 29 PCGACKFLRRKCVPGCIFAPYFDSEQgAAHFAAVHKVFGASNVSKLLLHIPVHKRLDAVVTICYEAQARLRDPVYGCVAHIFALQQQVVSLQAELTYLQA 128
7***********************9989********************************************************************9987 PP
DUF260 100 e 100
+
Cla006955 129 H 129
6 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50891 | 22.914 | 28 | 130 | IPR004883 | Lateral organ boundaries, LOB |
| Pfam | PF03195 | 1.1E-39 | 29 | 127 | IPR004883 | Lateral organ boundaries, LOB |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010089 | Biological Process | xylem development | ||||
| GO:0010311 | Biological Process | lateral root formation | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 240 aa Download sequence Send to blast |
MSANPSTSGG GGPGTASAGG GVGGGGGGPC GACKFLRRKC VPGCIFAPYF DSEQGAAHFA 60 AVHKVFGASN VSKLLLHIPV HKRLDAVVTI CYEAQARLRD PVYGCVAHIF ALQQQVVSLQ 120 AELTYLQAHL ATLELPSPPN PPPPPPPPSF PAQPPLSISD LPSASSLPAT YDLSSLFDPM 180 AQPAWAMQQQ RLDPRQFAGG GGGSSTAAGE GDLQALAREL LHRHGALPCS DASSPQSLSK |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5ly0_A | 3e-38 | 25 | 123 | 7 | 104 | LOB family transfactor Ramosa2.1 |
| 5ly0_B | 3e-38 | 25 | 123 | 7 | 104 | LOB family transfactor Ramosa2.1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the positive regulation of tracheary element (TE) differentiation. Involved in a positive feedback loop that maintains or promotes NAC030/VND7 expression that regulates TE differentiation-related genes (PubMed:19088331). Functions in the initiation and emergence of lateral roots, in conjunction with LBD16, downstream of ARF7 and ARF19 (PubMed:19717544, PubMed:23749813). Transcriptional activator that directly regulates EXPA14, a gene encoding a cell wall-loosening factor that promotes lateral root emergence. Activates EXPA14 by directly binding to a specific region of its promoter (PubMed:22974309). Transcriptional activator that directly regulates EXPA17, a gene encoding a cell wall-loosening factor that promotes lateral root emergence (PubMed:23872272). Acts downstream of the auxin influx carriers AUX1 and LAX1 in the regulation of lateral root initiation and development (PubMed:26059335). {ECO:0000269|PubMed:19088331, ECO:0000269|PubMed:19717544, ECO:0000269|PubMed:22974309, ECO:0000269|PubMed:23749813, ECO:0000269|PubMed:23872272, ECO:0000269|PubMed:26059335}. | |||||
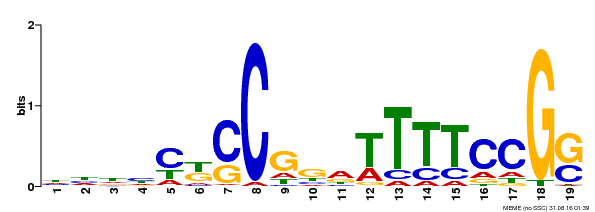
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00314 | DAP | Transfer from AT2G45420 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By auxin. {ECO:0000269|PubMed:15659631, ECO:0000269|PubMed:19717544, ECO:0000269|PubMed:23749813}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN684337 | 1e-151 | LN684337.1 Cucumis melo genomic scaffold, unassembled_sequence31840. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_023005988.1 | 1e-119 | LOB domain-containing protein 18-like | ||||
| Swissprot | O22131 | 3e-82 | LBD18_ARATH; LOB domain-containing protein 18 | ||||
| TrEMBL | A0A0A0KEN5 | 1e-117 | A0A0A0KEN5_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004145058.1 | 1e-118 | (Cucumis sativus) | ||||
| STRING | XP_004153747.1 | 1e-118 | (Cucumis sativus) | ||||
| STRING | XP_004170755.1 | 1e-118 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1771 | 34 | 93 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G45420.1 | 5e-77 | LOB domain-containing protein 18 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



