 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cla007663 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Citrullus
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 513aa MW: 55834.4 Da PI: 6.6087 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 52.5 | 1.1e-16 | 35 | 82 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT Ed +l+++vk++G g+W+++ ++ g+ R++k+c++rw ++l
Cla007663 35 KGPWTSAEDGILIEYVKKHGEGNWNAVQKHTGLARCGKSCRLRWANHL 82
79******************************************9986 PP
| |||||||
| 2 | Myb_DNA-binding | 49.8 | 7.8e-16 | 88 | 131 | 1 | 46 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqk 46
+g++++eE+ ++++++++lG++ W++ a+ ++ gRt++++k++w++
Cla007663 88 KGSFSQEEERIIIELHAKLGNK-WARMAAQLP-GRTDNEIKNYWNT 131
799*******************.*********.***********97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 16.604 | 30 | 82 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.42E-29 | 33 | 129 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 5.1E-13 | 34 | 84 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 2.7E-15 | 35 | 82 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 9.2E-24 | 36 | 89 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 3.66E-11 | 37 | 82 | No hit | No description |
| PROSITE profile | PS51294 | 25.983 | 83 | 137 | IPR017930 | Myb domain |
| SMART | SM00717 | 5.5E-16 | 87 | 135 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 9.9E-15 | 88 | 131 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.1E-25 | 90 | 136 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.70E-11 | 90 | 131 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009555 | Biological Process | pollen development | ||||
| GO:0009789 | Biological Process | positive regulation of abscisic acid-activated signaling pathway | ||||
| GO:0043068 | Biological Process | positive regulation of programmed cell death | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0090406 | Cellular Component | pollen tube | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 513 aa Download sequence Send to blast |
MVESKGEKGK SEGDHQHHAG SGGGGDNGGG RALKKGPWTS AEDGILIEYV KKHGEGNWNA 60 VQKHTGLARC GKSCRLRWAN HLRPNLKKGS FSQEEERIII ELHAKLGNKW ARMAAQLPGR 120 TDNEIKNYWN TRMKRRQRAG LPLYPLEIQQ EATAFHLRQH HHHHHHQPQQ QQHNSASAVA 180 TNFSAIRHKP DFNNHHPNSV SIFNFSSNMN NSQKNFNDGS SFYATPTSQF KFFPENNNGG 240 GGFPLPLSPV SPFPQIGQQM NQSFSPPPQA ALQLSYGGYV CNSNSGLNSM ILGAPYHHLI 300 PGLETELPSI QTPPHSTTPA SSGTSGGEGI MAAANSGLLD VVLLEAEARS RNEKQSKEES 360 SSAGEMKQRI NQGSTEDEDA TLYVESVLGS SGGETTAAAA ENHSDEFSSS HSSSRKRPRM 420 EPLEEMDSMD DDDLMSLLNN FQSGMPVPEW YPGSSNDDLG INMHNGPSLC ESNGNPGGDE 480 QRQNVASPSA VASSPVLEWS LGSSCWNNMP SIC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1a5j_A | 3e-33 | 33 | 136 | 5 | 107 | B-MYB |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00287 | DAP | Transfer from AT2G32460 | Download |
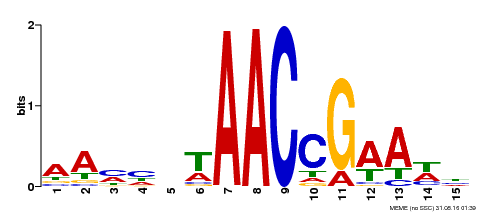
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681792 | 0.0 | LN681792.1 Cucumis melo genomic scaffold, anchoredscaffold00034. | |||
| GenBank | LN713255 | 0.0 | LN713255.1 Cucumis melo genomic chromosome, chr_1. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008455547.1 | 0.0 | PREDICTED: transcription factor MYB86 | ||||
| TrEMBL | A0A1S3C0R5 | 0.0 | A0A1S3C0R5_CUCME; transcription factor MYB86 | ||||
| STRING | XP_008455547.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF9949 | 32 | 40 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G32460.2 | 8e-72 | myb domain protein 101 | ||||



