 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cla009063 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Citrullus
|
||||||||
| Family | ERF | ||||||||
| Protein Properties | Length: 231aa MW: 25230.9 Da PI: 4.5349 | ||||||||
| Description | ERF family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 60.8 | 3.1e-19 | 26 | 74 | 3 | 55 |
AP2 3 ykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
+ GVr+++ +gr++AeIrdps+ + r++lg+f+taeeAa a+++a+++l+g
Cla009063 26 FLGVRRRP-WGRYAAEIRDPST---KERHWLGTFDTAEEAAIAYDRAARSLRG 74
67******.**********965...5*************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51032 | 20.468 | 25 | 82 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 5.0E-34 | 25 | 88 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 4.05E-29 | 25 | 81 | No hit | No description |
| PRINTS | PR00367 | 3.5E-8 | 26 | 37 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 3.99E-21 | 26 | 83 | IPR016177 | DNA-binding domain |
| Gene3D | G3DSA:3.30.730.10 | 2.5E-28 | 26 | 82 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 3.7E-11 | 28 | 74 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 3.5E-8 | 48 | 64 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0010030 | Biological Process | positive regulation of seed germination | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 231 aa Download sequence Send to blast |
MSPSPSKPKR KQQPPPPPAE PTPHRFLGVR RRPWGRYAAE IRDPSTKERH WLGTFDTAEE 60 AAIAYDRAAR SLRGSLARTN FLYSDSPPPP PPPSSASIYP DQPPLFLPLL QPPPPPLFHQ 120 FPPAGDSPSG IFSGECDGGA ELPPFPPAIS SESENCDYNY NYNYGLMESQ NVGFECFDQS 180 SIGIEACLGF DSSSEYVHNP MYGSMPAVSD AGAADFQSPA ANFYFPPSSD Y |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5wx9_A | 2e-18 | 25 | 81 | 14 | 71 | Ethylene-responsive transcription factor ERF096 |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00509 | DAP | Transfer from AT5G13910 | Download |
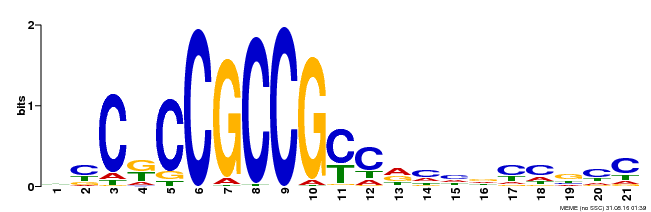
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004151396.1 | 1e-106 | PREDICTED: ethylene-responsive transcription factor LEP-like | ||||
| TrEMBL | A0A1S3AU89 | 1e-102 | A0A1S3AU89_CUCME; ethylene-responsive transcription factor LEP | ||||
| STRING | XP_004151396.1 | 1e-105 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3904 | 31 | 61 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G13910.1 | 1e-22 | ERF family protein | ||||



