 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cla009130 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Citrullus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 480aa MW: 53829.8 Da PI: 6.727 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.1 | 3e-16 | 69 | 113 | 1 | 47 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
r rW +eE+e++++a k++G + W+ I +++g ++t+ q++s+ qk+
Cla009130 69 RERWKEEEHEKFIEALKLYGRD-WRQIEEHVG-TKTAVQIRSHAQKF 113
78******************77.*********.************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 3.14E-15 | 63 | 118 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 21.519 | 64 | 118 | IPR017930 | Myb domain |
| TIGRFAMs | TIGR01557 | 5.3E-16 | 67 | 116 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 2.1E-13 | 68 | 116 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.7E-9 | 69 | 109 | IPR009057 | Homeodomain-like |
| Pfam | PF00249 | 2.6E-13 | 69 | 112 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.36E-10 | 71 | 114 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009734 | Biological Process | auxin-activated signaling pathway | ||||
| GO:0010600 | Biological Process | regulation of auxin biosynthetic process | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 480 aa Download sequence Send to blast |
MLYDRPMHHF LLANSGNINM NTGGDLCSND ISSAAYEPTV TPDQHSSGNA QLKEFSPKVR 60 KPYTITKQRE RWKEEEHEKF IEALKLYGRD WRQIEEHVGT KTAVQIRSHA QKFFSKVTRN 120 SNGCSTSSVG CIEIPPPRPK RKPAHPYPRK EVPHSHKASP ISELTRSLSP QLSEKECQSP 180 TSIVVSASGS DMLMFTDSRI HHDSGSPDSS IPSTEPNSSL LDNESPTAAP GTENSIPHEK 240 IPTNLELFTK DDMIEKDDST KEVSIQSLKL FGRTVLITDP HRQTPIIENC TPEADRKPGE 300 NQKQVSPWNS MVMESSTRNT ECTWNHGAFY FIQLNSGDSK QGPGSAVPVS WWSSYGSYPL 360 SFVHCFKQAD LDPNQVDDKE THKDQSWCGS NTGSVNSAEN GDKVGENDIQ SCRLFHNNRD 420 RDSVSKDELI GKALSSELIS SHEKNTKGFV PYKRCMTERA TQSHTITEAE REEKRIRLCL |
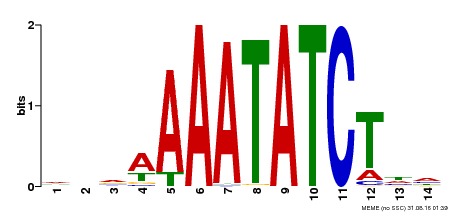
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00515 | DAP | Transfer from AT5G17300 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681894 | 0.0 | LN681894.1 Cucumis melo genomic scaffold, anchoredscaffold00074. | |||
| GenBank | LN713263 | 0.0 | LN713263.1 Cucumis melo genomic chromosome, chr_9. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011655141.1 | 0.0 | PREDICTED: protein REVEILLE 1-like isoform X1 | ||||
| TrEMBL | A0A0A0KPX5 | 0.0 | A0A0A0KPX5_CUCSA; Uncharacterized protein | ||||
| STRING | XP_008463161.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2956 | 33 | 72 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G17300.1 | 4e-52 | MYB_related family protein | ||||



