 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cla009916 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Citrullus
|
||||||||
| Family | C2H2 | ||||||||
| Protein Properties | Length: 310aa MW: 35813.9 Da PI: 8.1694 | ||||||||
| Description | C2H2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-C2H2 | 18.2 | 7.1e-06 | 22 | 42 | 3 | 23 |
ETTTTEEESSHHHHHHHHHHT CS
zf-C2H2 3 CpdCgksFsrksnLkrHirtH 23
C+ Cg F+ + +Lk+H+++H
Cla009916 22 CQECGAEFRKPAYLKQHMQSH 42
*******************99 PP
| |||||||
| 2 | zf-C2H2 | 16.1 | 3.3e-05 | 48 | 72 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
y C+ C s++rk++L+rH+ H
Cla009916 48 YICSvdGCEASYRRKDHLTRHLLQH 72
899999***************9887 PP
| |||||||
| 3 | zf-C2H2 | 21.9 | 4.6e-07 | 115 | 139 | 1 | 23 |
EEET..TTTEEESSHHHHHHHHHHT CS
zf-C2H2 1 ykCp..dCgksFsrksnLkrHirtH 23
++Cp Cgk+F+ s+L++H +H
Cla009916 115 FVCPedGCGKVFRYASQLQKHEDSH 139
89*******************9888 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS50157 | 12.944 | 20 | 47 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.015 | 20 | 42 | IPR015880 | Zinc finger, C2H2-like |
| Gene3D | G3DSA:3.30.160.60 | 2.5E-7 | 21 | 41 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE pattern | PS00028 | 0 | 22 | 42 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 3.24E-11 | 34 | 74 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 1.4E-12 | 42 | 74 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| PROSITE profile | PS50157 | 9.224 | 48 | 74 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 0.0018 | 48 | 72 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 50 | 72 | IPR007087 | Zinc finger, C2H2 |
| SuperFamily | SSF57667 | 7.14E-6 | 58 | 113 | No hit | No description |
| SMART | SM00355 | 0.17 | 77 | 102 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE profile | PS50157 | 12.133 | 77 | 107 | IPR007087 | Zinc finger, C2H2 |
| PROSITE pattern | PS00028 | 0 | 79 | 102 | IPR007087 | Zinc finger, C2H2 |
| Gene3D | G3DSA:3.30.160.60 | 6.7E-9 | 109 | 139 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SuperFamily | SSF57667 | 1.47E-5 | 112 | 140 | No hit | No description |
| PROSITE profile | PS50157 | 11.074 | 115 | 144 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 2.6E-4 | 115 | 139 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 117 | 139 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 1.5 | 147 | 172 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 149 | 172 | IPR007087 | Zinc finger, C2H2 |
| SMART | SM00355 | 4.8 | 175 | 197 | IPR015880 | Zinc finger, C2H2-like |
| SMART | SM00355 | 27 | 206 | 227 | IPR015880 | Zinc finger, C2H2-like |
| SuperFamily | SSF57667 | 3.78E-6 | 215 | 255 | No hit | No description |
| Gene3D | G3DSA:3.30.160.60 | 4.9E-7 | 217 | 255 | IPR013087 | Zinc finger C2H2-type/integrase DNA-binding domain |
| SMART | SM00355 | 12 | 233 | 259 | IPR015880 | Zinc finger, C2H2-like |
| PROSITE pattern | PS00028 | 0 | 235 | 259 | IPR007087 | Zinc finger, C2H2 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008097 | Molecular Function | 5S rRNA binding | ||||
| GO:0046872 | Molecular Function | metal ion binding | ||||
| GO:0080084 | Molecular Function | 5S rDNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 310 aa Download sequence Send to blast |
MEIGKHNDDG DDKTKNRKSN ICQECGAEFR KPAYLKQHMQ SHSLERPYIC SVDGCEASYR 60 RKDHLTRHLL QHQGIIFKCP IESCNREFAY ISCVNRHLKE CHGGEAPPEE CQKQFVCPED 120 GCGKVFRYAS QLQKHEDSHV KLDSIEACCT EPGCLRIFTN KQCLRAHMQS CHQYIKCEIC 180 GEKKLRKNLK QHLRQKHESE GPLEAIPCRY EGCSKASNLQ QHMKVVHMGQ KPFACCFPGC 240 GMRFGYKHVR DNHEKSGQHV YTNGDFEAAD EQFRSKPRGG RKRICPTVEM LIRKRVTPPS 300 EFDFTMDEEC |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5v3m_C | 1e-13 | 22 | 267 | 64 | 314 | Zinc finger protein 568 |
| 5wjq_D | 1e-13 | 22 | 263 | 39 | 285 | Zinc finger protein 568 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Essential protein (PubMed:22353599). Isoform 1 is a transcription activator the binds both 5S rDNA and 5S rRNA and stimulates the transcription of 5S rRNA gene (PubMed:12711688, PubMed:22353599). Isoform 1 regulates 5S rRNA levels during development (PubMed:22353599). {ECO:0000269|PubMed:12711688, ECO:0000269|PubMed:22353599}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00229 | DAP | Transfer from AT1G72050 | Download |
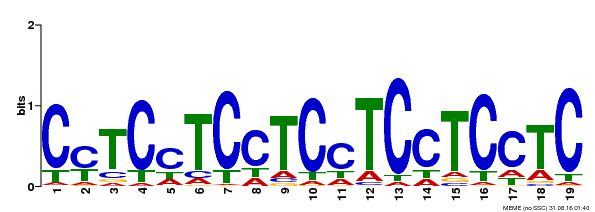
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | EU284120 | 1e-108 | EU284120.1 Cucumis sativus C2H2 zinc finger protein 1 mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004142387.2 | 0.0 | PREDICTED: zinc finger X-linked protein ZXDB | ||||
| Swissprot | Q84MZ4 | 1e-119 | TF3A_ARATH; Transcription factor IIIA | ||||
| TrEMBL | A0A0A0KV54 | 0.0 | A0A0A0KV54_CUCSA; Uncharacterized protein | ||||
| STRING | XP_008446968.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3069 | 33 | 70 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G72050.2 | 1e-121 | transcription factor IIIA | ||||



