 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cla012849 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Citrullus
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1098aa MW: 122453 Da PI: 5.4915 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 182.9 | 3.5e-57 | 16 | 132 | 2 | 118 |
CG-1 2 lkekkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfqrrcywl 101
++ k+rwl++ ei++iL n++k+++t+e+++rp+sgsl+L++rk++ryfrkDG++w+kkkdgktvrE+hekLKvg+++vl+cyYah+een++fqrr+yw+
Cla012849 16 IEAKHRWLRPAEICEILRNYSKFRITSEPPDRPSSGSLFLFDRKVLRYFRKDGHKWRKKKDGKTVREAHEKLKVGSIDVLHCYYAHGEENENFQRRSYWM 115
4569************************************************************************************************ PP
CG-1 102 Leeelekivlvhylevk 118
Lee+l +iv+vhylevk
Cla012849 116 LEEHLMHIVFVHYLEVK 132
**************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 82.756 | 11 | 137 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 6.0E-81 | 14 | 132 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 1.3E-50 | 17 | 131 | IPR005559 | CG-1 DNA-binding domain |
| Gene3D | G3DSA:2.60.40.10 | 9.9E-8 | 486 | 588 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF01833 | 2.8E-5 | 502 | 575 | IPR002909 | IPT domain |
| SuperFamily | SSF81296 | 5.29E-16 | 503 | 587 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 7.15E-16 | 683 | 800 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 3.1E-15 | 685 | 801 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.28E-12 | 688 | 798 | No hit | No description |
| PROSITE profile | PS50297 | 17.688 | 688 | 810 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50088 | 8.549 | 738 | 771 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 3.19E-8 | 920 | 971 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.0099 | 920 | 942 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 5.9E-4 | 922 | 941 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 8.261 | 922 | 950 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0038 | 943 | 965 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.633 | 944 | 968 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 2.3E-4 | 946 | 965 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1098 aa Download sequence Send to blast |
MADTAYILAD IEQLLIEAKH RWLRPAEICE ILRNYSKFRI TSEPPDRPSS GSLFLFDRKV 60 LRYFRKDGHK WRKKKDGKTV REAHEKLKVG SIDVLHCYYA HGEENENFQR RSYWMLEEHL 120 MHIVFVHYLE VKGNRTNAGA VVETDEVSSS SQKSSPRSSS YSSSHNQAAS ENADSPSPTS 180 TLTSFCEDAD AEDTYQATSR FHSFPTLPKM GNGLLVNKQD ADQSKFYFPH SSSNNLDAWS 240 SGPSVDYVTQ VQKAGLGSND GDTGVMGSQK TLSSASWEEI LQQCTTGFQT VPSHVLTSSI 300 EPLSSGIVLG QENSIPSKLL TSNSAIKEDF GSSLTMTSNW QVPFEDNTLS FSKEHADQYP 360 DLYSVYDIDS RFEQKSHDST FGSGHEMFSA HPGIENEDIL PNLELQFKEG ELYSAMRLSS 420 ENDMSKEGTI SYSLTLKQSL IDGEESLKKV DSFSRWVSKE LGEVDDLHMH PSSGLSWTTV 480 ECGDMVDDSS LSPSISEDQL FSITAFSPKW TLADLETEVV IIGKFLGNNH GTNCHWSCMF 540 GEVEVPAEVL ADGILCCHAP PHSVGRVPFY VTCSNRVACS EVREFDYLAG SAQDVDVTDI 600 YTVGSTKELC MHLRLERLLS LGSLDPSNDL SEDALEKRNL IRELITIKEE EDLYGEEPNS 660 QNDQIQHQNK DKEFLFVKLM KEKLYSWLIH KVIEGGKGPN ILDGDGQGVI HLAAALGYDW 720 AIRPIVAAGV SINFRDINGW TAVHWAASCG RQELTVGTLM SLDADPGLPS DPSPEFPWGR 780 TPADLASDNG HKGISGFLAE AALTSYVKSV SMADSISMGE AVEDGVADVS KRKPVHTVSE 840 RRATPVNDGF MPGDLSLKDS LTAVCNATQA AGRIHLMFRM QSFQRKKLSE CGTDEFGSSD 900 DSALSFVKAR ARKSGLSNSP VHAAAVQIQK KFRGWRMRKE FLLIRQRIVK IQAHVRGHQV 960 RKQYKKIVWS VGMIDKIILR WRRKGSGLRG FRADAVAKDP PSVMAPPTKE DDYDFLKEGR 1020 RQTEERFQKA LTRVKSMAQY PEGRDQYRRL LTVVQKCRET KGSHMAVTST SEEIIEGDDM 1080 IDIDTLLDDD ALMSMTFD |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
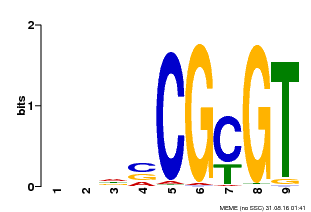
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681792 | 0.0 | LN681792.1 Cucumis melo genomic scaffold, anchoredscaffold00034. | |||
| GenBank | LN713255 | 0.0 | LN713255.1 Cucumis melo genomic chromosome, chr_1. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008455205.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 2 isoform X2 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | A0A1S3C137 | 0.0 | A0A1S3C137_CUCME; calmodulin-binding transcription activator 2 isoform X2 | ||||
| STRING | XP_008455204.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF7406 | 26 | 42 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



