 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cla013417 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Citrullus
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 591aa MW: 67251.4 Da PI: 7.0372 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 29.4 | 1.8e-09 | 540 | 588 | 2 | 47 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHT...TTS-HHHHHHHHHHH CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmg...kgRtlkqcksrwqky 47
++W+ E++ l av+++G+g+Wk+I + Rt+ ++k++w++
Cla013417 540 KKWSLLEEDTLRIAVQRFGKGNWKLILNSYRdifDERTEVDLKDKWRNM 588
58********************************99***********95 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 10.234 | 531 | 591 | IPR017930 | Myb domain |
| SMART | SM00717 | 5.5E-5 | 538 | 591 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.3E-8 | 540 | 588 | IPR001005 | SANT/Myb domain |
| CDD | cd11660 | 3.77E-17 | 540 | 590 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 4.2E-10 | 541 | 590 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 4.4E-13 | 541 | 590 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009901 | Biological Process | anther dehiscence | ||||
| GO:0010152 | Biological Process | pollen maturation | ||||
| GO:0043067 | Biological Process | regulation of programmed cell death | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 591 aa Download sequence Send to blast |
MDDDICRWII EFILRSPMDN HLQKRVMAVV PLSDKDFRLK KTVLLKSIES EILEVVLTEK 60 LLGIFELIEQ LDKAEGLALM ESMKAAYCAV AVECTVKFLL VEGICKHGRY FDAVKRIWRG 120 RVTELERSGK SELVSRELKA WKDEFEASLC DKNIRKKLMH VNTRYDALKL TGDYLSEVWA 180 VMGPSFIQLS ASLMDKRMVN GMQPIQLGQE TNKFASERED VGGSAGIELP SQTDNRAGPK 240 WQGRGGVLSQ PRTRTDLPNS HQDLGTNEDS KQPAIVAMTT ERVQELATET AEGQESVEKE 300 VAVLHNPSPH RDERVQELAT ETAEGQESVE KEVAVLHNPS PRQDENVRTS AMPRCKSLAF 360 HRRVRGARLS QLEDLENEND NSFERYTRLD TPEVNRVREA LRTSSLELQA VVKDPLPDAL 420 RIAESVANDL AERNKTCENP LEGRNDAGAT NPTINKGVLP LQSVSANLKN PGYGHKTVFP 480 RPSIMERNST ACTYEWNDSI DDSPEGSHAN RLRLPSPKRK VISPLKKYEE TKVVRRRQCK 540 KWSLLEEDTL RIAVQRFGKG NWKLILNSYR DIFDERTEVD LKDKWRNMTR Y |
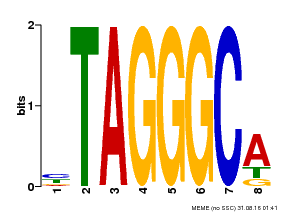
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00052 | PBM | Transfer from AT1G15720 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681920 | 0.0 | LN681920.1 Cucumis melo genomic scaffold, anchoredscaffold00113. | |||
| GenBank | LN713265 | 0.0 | LN713265.1 Cucumis melo genomic chromosome, chr_11. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008465305.1 | 0.0 | PREDICTED: uncharacterized protein LOC103502960 isoform X1 | ||||
| TrEMBL | A0A1S3CNK2 | 0.0 | A0A1S3CNK2_CUCME; uncharacterized protein LOC103502960 isoform X1 | ||||
| STRING | XP_008465305.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2824 | 32 | 60 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G15720.1 | 1e-21 | TRF-like 5 | ||||



