 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cla017743 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Citrullus
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 200aa MW: 22634.5 Da PI: 7.5674 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 62.6 | 5.9e-20 | 26 | 79 | 3 | 56 |
--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 3 kRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
k+ ++++eqle Le+ F+++ +++ +++++L+k+lgL+ rq+ vWFqNrRa++k
Cla017743 26 KKKRLSSEQLESLERSFQEEIKLDPDRKQKLSKELGLQPRQIAVWFQNRRARWK 79
45679************************************************9 PP
| |||||||
| 2 | HD-ZIP_I/II | 120.3 | 1e-38 | 25 | 117 | 1 | 93 |
HD-ZIP_I/II 1 ekkrrlskeqvklLEesFeeeekLeperKvelareLglqprqvavWFqnrRARtktkqlEkdyeaLkraydalkeenerLekeveeLreelke 93
ekk+rls+eq+++LE+sF+ee kL+p+rK++l++eLglqprq+avWFqnrRAR+k+kqlE+ y++Lk+++da+++e+++L++ev +L++ l+e
Cla017743 25 EKKKRLSSEQLESLERSFQEEIKLDPDRKQKLSKELGLQPRQIAVWFQNRRARWKAKQLEHLYDTLKQEFDAISREKHKLQEEVMKLKSMLRE 117
69**************************************************************************************99876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF46689 | 3.29E-19 | 15 | 83 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.167 | 21 | 81 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.6E-17 | 23 | 85 | IPR001356 | Homeobox domain |
| Pfam | PF00046 | 2.9E-17 | 26 | 79 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 1.22E-16 | 26 | 82 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 4.0E-21 | 28 | 88 | IPR009057 | Homeodomain-like |
| PRINTS | PR00031 | 2.0E-5 | 52 | 61 | IPR000047 | Helix-turn-helix motif |
| PROSITE pattern | PS00027 | 0 | 56 | 79 | IPR017970 | Homeobox, conserved site |
| PRINTS | PR00031 | 2.0E-5 | 61 | 77 | IPR000047 | Helix-turn-helix motif |
| Pfam | PF02183 | 1.3E-8 | 81 | 115 | IPR003106 | Leucine zipper, homeobox-associated |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009965 | Biological Process | leaf morphogenesis | ||||
| GO:0010434 | Biological Process | bract formation | ||||
| GO:0010582 | Biological Process | floral meristem determinacy | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048510 | Biological Process | regulation of timing of transition from vegetative to reproductive phase | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 200 aa Download sequence Send to blast |
MAGASETVLQ GLVPGMDMNS YGNLEKKKRL SSEQLESLER SFQEEIKLDP DRKQKLSKEL 60 GLQPRQIAVW FQNRRARWKA KQLEHLYDTL KQEFDAISRE KHKLQEEVMK LKSMLRELQA 120 ARNQVSTVYT DLSGEETVES TSVGAGCSSK PRPVAAAAAV AAANHYPTPP EQCNYVFNTE 180 DYNPMSPTFW GSLPSSYHPQ |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 73 | 81 | RRARWKAKQ |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Putative transcription factor. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00486 | DAP | Transfer from AT5G03790 | Download |
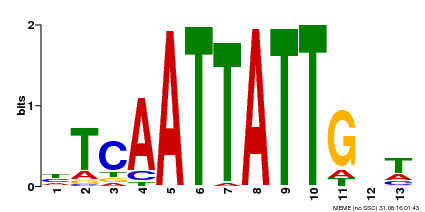
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KM095652 | 0.0 | KM095652.1 Cucumis sativus multicellular trichome development class I homeodomain-leucine zipper transcription factor (Gl) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008442780.1 | 1e-146 | PREDICTED: homeobox-leucine zipper protein ATHB-22-like isoform X1 | ||||
| Refseq | XP_011651984.1 | 1e-146 | PREDICTED: homeobox-leucine zipper protein ATHB-22-like isoform X1 | ||||
| Swissprot | Q9LZR0 | 4e-49 | ATB51_ARATH; Putative homeobox-leucine zipper protein ATHB-51 | ||||
| TrEMBL | A0A097A1J6 | 1e-144 | A0A097A1J6_CUCSA; Multicellular trichome development class I homeodomain-leucine zipper transcription factor | ||||
| TrEMBL | A0A1S3B6J1 | 1e-144 | A0A1S3B6J1_CUCME; homeobox-leucine zipper protein ATHB-22-like isoform X1 | ||||
| STRING | XP_008442780.1 | 1e-145 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3394 | 34 | 66 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G03790.1 | 2e-39 | homeobox 51 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



