 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cla018652 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Citrullus
|
||||||||
| Family | HD-ZIP | ||||||||
| Protein Properties | Length: 705aa MW: 78489.9 Da PI: 6.8396 | ||||||||
| Description | HD-ZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Homeobox | 61.3 | 1.4e-19 | 21 | 76 | 1 | 56 |
TT--SS--HHHHHHHHHHHHHSSS--HHHHHHHHHHCTS-HHHHHHHHHHHHHHHH CS
Homeobox 1 rrkRttftkeqleeLeelFeknrypsaeereeLAkklgLterqVkvWFqNrRakek 56
+++ ++++++q+++Le++F+++++p++++r +L+++lgL rq+k+WFqNrR+++k
Cla018652 21 KKRYHRHNANQIQRLEAMFKECPHPDEKQRLQLSRELGLAPRQIKFWFQNRRTQMK 76
6778999**********************************************998 PP
| |||||||
| 2 | START | 183.8 | 9.5e-58 | 219 | 443 | 2 | 206 |
HHHHHHHHHHHHHHHC-TT-EEEE....EXCCTTEEEEEEESSS.......SCEEEEEEEECCSCHHHHHHHHHCCCGGCT-TT-S....EEEEEEEECT CS
START 2 laeeaaqelvkkalaeepgWvkss....esengdevlqkfeeskv......dsgealrasgvvdmvlallveellddkeqWdetla....kaetleviss 87
+a a++el+++ + +ep+W+ks+ + ++ +++ q f++ ++ +++ea+r+sgvv+m +a lv +++d++ +W+e ++ a t+eviss
Cla018652 219 IATNAMAELLRLSQTNEPFWMKSPtdgrDLLDLETYEQAFPRPNTplknlhFRTEASRDSGVVIMSSAALVDIFMDSN-KWTELFPtivsIARTIEVISS 317
77899********************999999999999***999889********************************.*******999999******** PP
T......EEEEEEEEXXTTXX-SSX.EEEEEEEEEEE.TTS-EEEEEEEEE-TTS--.-TTSEE-EESSEEEEEEEECTCEEEEEEEE-EE--SSXX.HH CS
START 88 g......galqlmvaelqalsplvp.RdfvfvRyirqlgagdwvivdvSvdseqkppesssvvRaellpSgiliepksnghskvtwvehvdlkgrlp.hw 179
g g lqlm++elq+lsplvp R f+++Ry++q ++g+w++vdvS + +++++ s ++++pSg+li++++ng+skvtw+ehv++++r hw
Cla018652 318 GmlgsqnGSLQLMYQELQLLSPLVPtRHFYVLRYCQQIEQGVWAVVDVSYNIPRENQI-VSHSQCHRFPSGCLIQDMPNGYSKVTWIEHVEVEDRGStHW 416
********************************************************95.778889******************************999** PP
HHHHHHHHHHHHHHHHHHHHTXXXXXX CS
START 180 llrslvksglaegaktwvatlqrqcek 206
l+r l++sgla+ga +w+atlqr e+
Cla018652 417 LFRDLIHSGLAFGAERWLATLQRMAER 443
**********************98875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 4.9E-22 | 6 | 72 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 9.19E-20 | 7 | 78 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS50071 | 17.767 | 18 | 78 | IPR001356 | Homeobox domain |
| SMART | SM00389 | 1.5E-18 | 19 | 82 | IPR001356 | Homeobox domain |
| CDD | cd00086 | 4.25E-18 | 20 | 79 | No hit | No description |
| Pfam | PF00046 | 3.6E-17 | 21 | 76 | IPR001356 | Homeobox domain |
| PROSITE pattern | PS00027 | 0 | 53 | 76 | IPR017970 | Homeobox, conserved site |
| PROSITE profile | PS50848 | 47.171 | 209 | 446 | IPR002913 | START domain |
| SuperFamily | SSF55961 | 5.91E-36 | 212 | 444 | No hit | No description |
| CDD | cd08875 | 3.99E-114 | 213 | 442 | No hit | No description |
| SMART | SM00234 | 2.0E-50 | 218 | 443 | IPR002913 | START domain |
| Pfam | PF01852 | 2.5E-49 | 219 | 442 | IPR002913 | START domain |
| Gene3D | G3DSA:3.30.530.20 | 3.2E-7 | 278 | 442 | IPR023393 | START-like domain |
| SuperFamily | SSF55961 | 1.65E-22 | 464 | 697 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009828 | Biological Process | plant-type cell wall loosening | ||||
| GO:0010091 | Biological Process | trichome branching | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0000976 | Molecular Function | transcription regulatory region sequence-specific DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0008289 | Molecular Function | lipid binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 705 aa Download sequence Send to blast |
MDFGGGSGWD NDSSSDPQRR KKRYHRHNAN QIQRLEAMFK ECPHPDEKQR LQLSRELGLA 60 PRQIKFWFQN RRTQMKAQHE RADNSALRAE NDKIRCENIA IREALKNVIC PSCGGPPLQD 120 PYFDEHKLRI ENSHLKEELD RLSSIAAKYI GRPISQLPPI QPSHFSSLEL SMGSFPSQEM 180 GCPSLDLDLL PASSTSVTSL PYLHPIHLSS VDKSLMTEIA TNAMAELLRL SQTNEPFWMK 240 SPTDGRDLLD LETYEQAFPR PNTPLKNLHF RTEASRDSGV VIMSSAALVD IFMDSNKWTE 300 LFPTIVSIAR TIEVISSGML GSQNGSLQLM YQELQLLSPL VPTRHFYVLR YCQQIEQGVW 360 AVVDVSYNIP RENQIVSHSQ CHRFPSGCLI QDMPNGYSKV TWIEHVEVED RGSTHWLFRD 420 LIHSGLAFGA ERWLATLQRM AERFACLMVT GSSNQDLSGV IPSPEGKRSM MKLAQRMVNN 480 FCASISTSHG HRWTTLSGMN EVGVRVTVHK STDSGQPNGV VLSAATTIWL PVYPQTIFNF 540 FKNDRTRSQW DVLSDGNPVQ EVAHISNGSH PGNCISVLRA FNTSQNNMLI LQESCIDSSG 600 SLVVYCPVDL PAMNVAMSGE DPSSIPLLPS GFTILPDGRR DQGEGTSSSS DVNNRSGGSL 660 VTVAFQILVS SLPSGKLNLE SVTTVNNLIS TTVHQIKTAL NCHSS |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor which acts as positive regulator of drought stress tolerance. Can transactivate CIPK3, NCED3 and ERECTA (PubMed:18451323). Transactivates several cell-wall-loosening protein genes by directly binding to HD motifs in their promoters. These target genes play important roles in coordinating cell-wall extensibility with root development and growth (PubMed:24821957). Transactivates CYP74A/AOS, AOC3, OPR3 and 4CLL5/OPCL1 genes by directly binding to HD motifs in their promoters. These target genes are involved in jasmonate (JA) biosynthesis, and JA signaling affects root architecture by activating auxin signaling, which promotes lateral root formation (PubMed:25752924). Acts as negative regulator of trichome branching (PubMed:16778018, PubMed:24824485). Required for the establishment of giant cell identity on the abaxial side of sepals (PubMed:23095885). May regulate cell differentiation and proliferation during root and shoot meristem development (PubMed:25564655). {ECO:0000269|PubMed:16778018, ECO:0000269|PubMed:18451323, ECO:0000269|PubMed:23095885, ECO:0000269|PubMed:24821957, ECO:0000269|PubMed:24824485, ECO:0000269|PubMed:25564655, ECO:0000269|PubMed:25752924}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00232 | DAP | Transfer from AT1G73360 | Download |
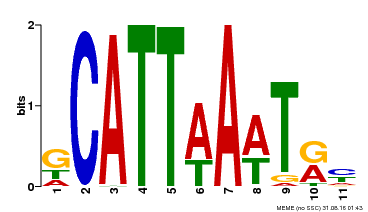
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681932 | 0.0 | LN681932.1 Cucumis melo genomic scaffold, anchoredscaffold00001. | |||
| GenBank | LN713266 | 0.0 | LN713266.1 Cucumis melo genomic chromosome, chr_12. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_016898817.1 | 0.0 | PREDICTED: homeobox-leucine zipper protein HDG11-like | ||||
| Swissprot | Q9FX31 | 0.0 | HDG11_ARATH; Homeobox-leucine zipper protein HDG11 | ||||
| TrEMBL | A0A1S4DS38 | 0.0 | A0A1S4DS38_CUCME; homeobox-leucine zipper protein HDG11-like | ||||
| STRING | XP_008445879.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1026 | 33 | 98 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G73360.1 | 0.0 | homeodomain GLABROUS 11 | ||||



