 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cla019641 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Citrullus
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 327aa MW: 35190.3 Da PI: 8.4856 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 209 | 1.3e-64 | 2 | 147 | 1 | 150 |
DUF822 1 ggsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeaeaagssasaspesslqsslks 100
+++ r+ptwkErEnnkrRERrRRaiaaki+ GLR++Gnyklpk++DnneVlkALc eAGw+ve+DGttyrkg+kp e +++g sasasp+ss+q
Cla019641 2 TSGSRMPTWKERENNKRRERRRRAIAAKIFLGLRMYGNYKLPKHCDNNEVLKALCDEAGWTVEEDGTTYRKGCKPV-ERDIMGGSASASPCSSYQ----- 95
5789************************************************************************.99****************..... PP
DUF822 101 salaspvesysaspksssfpspssldsislasa.....asllpvlsvlslvsssl 150
+sp++sy++sp+sssfpsp+s+ + + ++ +sl+p+l++ls+ sss+
Cla019641 96 ---PSPRASYNPSPASSSFPSPTSSRYAINGNDnatdpNSLIPWLKNLSSGSSSA 147
...*******************9977654333348889**********9977764 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 1.1E-65 | 4 | 141 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 327 aa Download sequence Send to blast |
MTSGSRMPTW KERENNKRRE RRRRAIAAKI FLGLRMYGNY KLPKHCDNNE VLKALCDEAG 60 WTVEEDGTTY RKGCKPVERD IMGGSASASP CSSYQPSPRA SYNPSPASSS FPSPTSSRYA 120 INGNDNATDP NSLIPWLKNL SSGSSSASSR LPHHLFITCG SISAPVTPPS SSPTARTPRK 180 PNDWDNHPAV APAWAAQRFS CLPASTPPSP GRQVLADPAW LDSMRIPQSG PSSPTFSLVA 240 RNPFGFKEAM SAGGSRNWTP AQSGTCSPTV AAGTDHTIDV PMMDGTAAEF AFGSCSVGIV 300 KPWEGERIHE ECISDDLELT LGNSSTR |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5zd4_A | 1e-24 | 6 | 86 | 372 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_B | 1e-24 | 6 | 86 | 372 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_C | 1e-24 | 6 | 86 | 372 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| 5zd4_D | 1e-24 | 6 | 86 | 372 | 452 | Maltose-binding periplasmic protein,Protein BRASSINAZOLE-RESISTANT 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00248 | DAP | Transfer from AT1G78700 | Download |
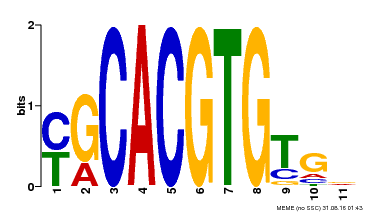
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681932 | 0.0 | LN681932.1 Cucumis melo genomic scaffold, anchoredscaffold00001. | |||
| GenBank | LN713266 | 0.0 | LN713266.1 Cucumis melo genomic chromosome, chr_12. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004147390.1 | 0.0 | PREDICTED: BES1/BZR1 homolog protein 4-like | ||||
| Swissprot | Q9ZV88 | 1e-121 | BEH4_ARATH; BES1/BZR1 homolog protein 4 | ||||
| TrEMBL | A0A0A0M073 | 0.0 | A0A0A0M073_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004172293.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2661 | 33 | 75 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G78700.1 | 1e-92 | BES1/BZR1 homolog 4 | ||||



