 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cla021770 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Citrullus
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 661aa MW: 72853.7 Da PI: 6.208 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 91.4 | 7.9e-29 | 204 | 257 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kpr++W+ eLH++Fv av+qL G++kA+Pk+ilelm+v+gLt+e+v+SHLQkYRl
Cla021770 204 KPRVVWSVELHQQFVAAVNQL-GIDKAVPKKILELMNVPGLTRENVASHLQKYRL 257
79*******************.********************************8 PP
| |||||||
| 2 | Response_reg | 76 | 1.4e-25 | 26 | 134 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTTES CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaGak 95
vl+vdD+p+ +++l+++l+ +y ev+ ++ +e+al ll++ + +D++l D+ mp+mdG++ll+ i e +lp+i+++ + ++ + + + Ga
Cla021770 26 VLVVDDDPTCLMILEKMLRICRY-EVTNCSRAEDALSLLRQYKngFDIVLSDVHMPDMDGFKLLEYIGLEM-DLPVIMMSVDDGKNVVMKGVTHGAC 120
89*********************.***************999999**********************6654.8************************ PP
EEEESS--HHHHHH CS
Response_reg 96 dflsKpfdpeelvk 109
d+l Kp+ +e+l +
Cla021770 121 DYLIKPVRIEALKN 134
**********9986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF036392 | 3.1E-175 | 5 | 651 | IPR017053 | Response regulator B-type, plant |
| Gene3D | G3DSA:3.40.50.2300 | 2.9E-40 | 23 | 165 | No hit | No description |
| SuperFamily | SSF52172 | 9.82E-35 | 23 | 147 | IPR011006 | CheY-like superfamily |
| SMART | SM00448 | 8.8E-28 | 24 | 136 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 40.741 | 25 | 140 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 1.3E-22 | 26 | 134 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 5.18E-25 | 27 | 139 | No hit | No description |
| SuperFamily | SSF46689 | 3.58E-20 | 201 | 261 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 11.048 | 201 | 260 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.4E-30 | 203 | 262 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 9.4E-25 | 204 | 257 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 9.4E-8 | 206 | 256 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009873 | Biological Process | ethylene-activated signaling pathway | ||||
| GO:0010082 | Biological Process | regulation of root meristem growth | ||||
| GO:0010119 | Biological Process | regulation of stomatal movement | ||||
| GO:0010150 | Biological Process | leaf senescence | ||||
| GO:0071368 | Biological Process | cellular response to cytokinin stimulus | ||||
| GO:0080113 | Biological Process | regulation of seed growth | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 661 aa Download sequence Send to blast |
MSTASSSSVR KAGEAVPDQF PAGLRVLVVD DDPTCLMILE KMLRICRYEV TNCSRAEDAL 60 SLLRQYKNGF DIVLSDVHMP DMDGFKLLEY IGLEMDLPVI MMSVDDGKNV VMKGVTHGAC 120 DYLIKPVRIE ALKNIWQHVV RKRKNEWKDL EQSGCVDDAD RQQKTNEDAD YSSSANEGSW 180 RNSKRRKDDV EDPEERDDSS TLKKPRVVWS VELHQQFVAA VNQLGIDKAV PKKILELMNV 240 PGLTRENVAS HLQKYRLYLR RLSGITQHQS NLNNTFMSAQ DAFVPSMNGL DLQTLAAAGQ 300 LQPQSLATLQ AAGFGRSTAK SGMPMPLVDQ RNHIFSFENP KLRFGDGQQP HLNGNKPMNL 360 LHGIPTTMEP KQLANLQHSA QSHGNMTMQV SAQGSQSSSQ LMQTPQPQAR AQILNESTST 420 SVTRLPPTMQ QSILPNGTTG AVLARNEFAN NSRGGGYNLV SPASTMLNFP LNQTAELPGN 480 SFPLQSTPGM SSIVPKGRFP DEVNSDIKGS EGFVSSYDMF RDLHPQKPHD WDLQHVGLTF 540 DTSQGSLDIP PSAFAHQGYA SSQQNGQNRN ASTAGKAMFL LEEGSDNGNA QSVGHQLNPL 600 FVDGSVRVKA ERAPDTSNQT DLFSDHFGQE DLMSALLKQQ QGSIATAESE LEWVFHHNTP 660 V |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 6e-22 | 203 | 263 | 4 | 64 | ARR10-B |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-[AG]GATT-3'. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. Could directly activate some type-A response regulators in response to cytokinins. Involved in the expression of nuclear genes for components of mitochondrial complex I. Promotes cytokinin-mediated leaf longevity. Involved in the ethylene signaling pathway in an ETR1-dependent manner and in the cytokinin signaling pathway. {ECO:0000269|PubMed:11370868, ECO:0000269|PubMed:11574878, ECO:0000269|PubMed:15282545, ECO:0000269|PubMed:16407152}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00054 | PBM | Transfer from AT4G16110 | Download |
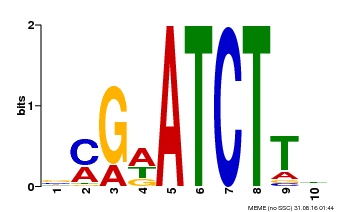
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681848 | 0.0 | LN681848.1 Cucumis melo genomic scaffold, anchoredscaffold00006. | |||
| GenBank | LN713260 | 0.0 | LN713260.1 Cucumis melo genomic chromosome, chr_6. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_008438918.1 | 0.0 | PREDICTED: two-component response regulator ARR2-like | ||||
| Swissprot | Q9ZWJ9 | 0.0 | ARR2_ARATH; Two-component response regulator ARR2 | ||||
| TrEMBL | A0A1S3AX53 | 0.0 | A0A1S3AX53_CUCME; Two-component response regulator | ||||
| STRING | XP_008438918.1 | 0.0 | (Cucumis melo) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF3958 | 34 | 58 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16110.1 | 0.0 | response regulator 2 | ||||



