 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cotton_A_12693_BGI-A2_v1.0 | ||||||||
| Common Name | F383_24542 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1066aa MW: 117636 Da PI: 8.2899 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 131.1 | 3.9e-41 | 151 | 228 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C++dls+ak+yhrrhkvCe+hska+++lv++++qrfCqqCsrfh lsefDe+krsCrrrLa+hn+rrrk+q+
Cotton_A_12693_BGI-A2_v1.0 151 MCQVDNCKEDLSNAKDYHRRHKVCEIHSKATKALVEKQMQRFCQQCSRFHPLSEFDEGKRSCRRRLAGHNRRRRKTQP 228
6**************************************************************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 8.2E-34 | 146 | 213 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.035 | 149 | 226 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.7E-37 | 150 | 230 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 4.9E-30 | 152 | 225 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 3.1E-4 | 814 | 951 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 7.83E-7 | 814 | 956 | No hit | No description |
| SuperFamily | SSF48403 | 3.32E-6 | 856 | 980 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1066 aa Download sequence Send to blast |
MEDAGAQVAP PLYIHQALAS RFCDPPSLPR KRDLSYQASD FLYQNPSQQR VANPRDNWNP 60 KQWEWDAVRF IAKPLNTEIL QTGTATAEQR KTGHVNGNEN SITSKKATAA NDDDERLQLN 120 LGGGLNSVED PVSRPNKKVC GGSPGSTSFP MCQVDNCKED LSNAKDYHRR HKVCEIHSKA 180 TKALVEKQMQ RFCQQCSRFH PLSEFDEGKR SCRRRLAGHN RRRRKTQPED VTSRLLLPAN 240 RDNAGNGSKT EDKSINPSPV PNRDQLLQIL SKINSLPLPM DLAAKLPNVG VLNRKSQEQP 300 SLGNQNQLNG KNTSSPSTVD LLAALSATLT SSSSDALAML SQRSSQSSDS QKTKSICPDN 360 VAASSSLNRA PLEFTSVGGE RSSTSYQSPV EDSECQIQET RANLPLQLFS SSPEDDSPPK 420 LASSRKYFSS DSSNPMEERS PSSSPVVQKF FPMHSTPEAV KYEKVPIGRH ANTNAETSRA 480 HGSIIPLELF SGSKRGTGRG SFQHFPSQVG YTSSSGSDHS PSSLNSDAQD RTGRIIFKLF 540 DKDPSHFPGT LRTQIYNWLS NSPSEMESYI RPGCVVLSVY VSMPAPAWEQ LEGNLLQYVN 600 CLLQDSDSDF WRKARFLVHT GNRQLASHKD GKIHLCKSWL SSSSPELISV SPLAVVSGQE 660 TSLLLRGRNL TNPGTEIHCA YMGGYSSMQI NGSTYKGASY DEANMGSFKI QVPSPKALGR 720 CFIEAENGFK GNSFPIIIAD AAICKELRLL ESELDTEVKA SDIISEEHAY DSHRPRSREE 780 VLHFLNELGW LFQRSTAPLP KSSDHSLRRF KFLLMFSVES DYCALVKVLL DMLVESNLDM 840 DDLSKDLLAM LSEIQLLTRA VKRRCRKMAD LLIHYSISSN DGNSKKYIFP PNLEGAGGIS 900 PLHLAACTSG SDDMVDVLTN DPQEIGLTCW SSLLDANGQS PYAYAMMRNN HSYNKLVAGK 960 YADRRNGQVS LTIGVEDQHS GVSAVQLNKI SLRFKQDRSS CAKCAVVATR SNNRFPGSQG 1020 LLQRPYVHSM LAIAAVCVCV CLFLRGSPNI GRVSPFKWEN LDFGTI |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 1e-32 | 142 | 225 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 858 | 863 | RAVKRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
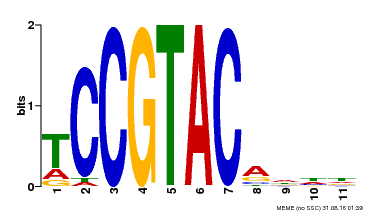
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KJ622322 | 0.0 | KJ622322.1 Gossypium hirsutum SQUAMOSA promoter binding-like transcription factor (SPL14) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017641665.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 14 | ||||
| Refseq | XP_017641666.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | A0A0B0P4N4 | 0.0 | A0A0B0P4N4_GOSAR; Squamosa promoter-binding-like protein 14 | ||||
| STRING | Gorai.002G112200.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3999 | 25 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



