 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cotton_A_13575_BGI-A2_v1.0 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | BES1 | ||||||||
| Protein Properties | Length: 706aa MW: 79934.6 Da PI: 5.8233 | ||||||||
| Description | BES1 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF822 | 138.9 | 5.2e-43 | 50 | 128 | 2 | 80 |
DUF822 2 gsgrkptwkErEnnkrRERrRRaiaakiyaGLRaqGnyklpkraDnneVlkALcreAGwvvedDGttyrkgskpleeae 80
g+g+++++kE+E++k+RER+RRai+++++aGLR++Gn++lp+raD+n+Vl+AL+reAGw+ve+DGttyr++ p+++++
Cotton_A_13575_BGI-A2_v1.0 50 GKGKREREKEKERTKLRERHRRAITSRMLAGLRQYGNFPLPARADMNDVLAALAREAGWTVEPDGTTYRHSPPPQHQQH 128
899******************************************************************8777773333 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05687 | 2.0E-42 | 50 | 132 | IPR008540 | BES1/BZR1 plant transcription factor, N-terminal |
| SuperFamily | SSF51445 | 9.18E-156 | 264 | 700 | IPR017853 | Glycoside hydrolase superfamily |
| Gene3D | G3DSA:3.20.20.80 | 8.1E-169 | 267 | 698 | IPR013781 | Glycoside hydrolase, catalytic domain |
| Pfam | PF01373 | 1.4E-81 | 291 | 661 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.3E-54 | 304 | 318 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.3E-54 | 325 | 343 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.3E-54 | 347 | 368 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.3E-54 | 440 | 462 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.3E-54 | 513 | 532 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.3E-54 | 547 | 563 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.3E-54 | 564 | 575 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.3E-54 | 582 | 605 | IPR001554 | Glycoside hydrolase, family 14 |
| PRINTS | PR00750 | 1.3E-54 | 620 | 642 | IPR001554 | Glycoside hydrolase, family 14 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000272 | Biological Process | polysaccharide catabolic process | ||||
| GO:0048831 | Biological Process | regulation of shoot system development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0016161 | Molecular Function | beta-amylase activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 706 aa Download sequence Send to blast |
MESQPQPQPQ PQPQPRRPRG FAATAAAAAA SVSPTGAPSG SAGASTASSG KGKREREKEK 60 ERTKLRERHR RAITSRMLAG LRQYGNFPLP ARADMNDVLA ALAREAGWTV EPDGTTYRHS 120 PPPQHQQHLY ILCSSQYPDN FSGHPYCKCF TGCYFMSTVL DLSLWQPYFD IDNAQGAFSV 180 RSGESPLSAT SLKNCSVKAT LDCQQPVVRI DDSLSPASLD SVVIAERDTR SEKYPSTSPI 240 NSVECLEADQ LIQDVHSTEH DNDFTGSQYV PVYVKLSTGV INNFCQLADP DDVRQELSHM 300 NSLNVDGVIV DCWWGIVECW NPQKYVWSGY RELFNYIREF KMKIQVVMAF HEYGRTDSAD 360 VLISLPNWIL EIGKENQDIF FTDREGRRTT EFLSWGIDKE RVLNGRTGVE VYFDLMRSFR 420 TEFDDLFAEG LISAVEIGLG PSGELRYPSF SERMGWRYPG IGEFQCYDKY LQQHLRKAAK 480 LRGHSFWARG PDNAGHYNSR PHETGFFCER GDYDSYYGRF FLHWYAQALM DHADNVLSLA 540 NLAFEETKII VKIPAVYWWY KTSSHAAELT AGYNNPTNQD GYSPVFEVLK KHSVTVKFVC 600 YGLQVCSYEN DEAFADPEGL SWQVLNSAWD RGLKVAGENT LSCYDREGCL RIIETAKPRN 660 DPDRRHFSFF VYQQPSPLVQ GVICLPDLDY FIKCMHGDIT GDLVPS |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wdq_A | 1e-121 | 268 | 659 | 10 | 401 | Beta-amylase |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00543 | DAP | Transfer from AT5G45300 | Download |
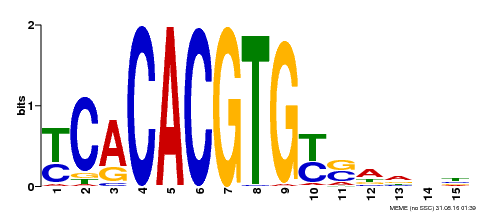
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017637364.1 | 0.0 | PREDICTED: beta-amylase 8 isoform X2 | ||||
| Swissprot | Q9FH80 | 0.0 | BAM8_ARATH; Beta-amylase 8 | ||||
| TrEMBL | A0A1U8N730 | 0.0 | A0A1U8N730_GOSHI; Beta-amylase | ||||
| STRING | Gorai.008G218300.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM10888 | 28 | 32 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45300.2 | 0.0 | beta-amylase 2 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



