 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cotton_A_15020_BGI-A2_v1.0 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 394aa MW: 43825.1 Da PI: 6.8791 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 101.3 | 6.1e-32 | 34 | 88 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kprl+Wtp+LHe F+ea +qLGG++kAtPkt+++lm+++gLtl+h+kSHLQkYRl
Cotton_A_15020_BGI-A2_v1.0 34 KPRLKWTPDLHEXFIEAANQLGGADKATPKTVMKLMGIPGLTLYHLKSHLQKYRL 88
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 9.454 | 31 | 91 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 8.1E-30 | 32 | 89 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 9.5E-16 | 33 | 89 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.3E-21 | 34 | 89 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.6E-8 | 36 | 87 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 3.0E-24 | 143 | 189 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 394 aa Download sequence Send to blast |
YQHQGKNIHP ERHLFLQGGN GPGDSGLVLS TDAKPRLKWT PDLHEXFIEA ANQLGGADKA 60 TPKTVMKLMG IPGLTLYHLK SHLQKYRLSK NLHGQANNGS NKIGKMTVFP FSAVAMAGDR 120 MSEATGTHMN NLSIGPQTNK GLQIGEALQM QIEVQRRLHE QLEVQRHLQL RIEAQGKYLQ 180 AVLEKAQETL GRQNLGNMGL EAAKVQLSEL VSKVSNQCLN SAFPDLKDLQ GLCPQQGQTQ 240 ATPPTDCSMD SCLTSCDGSQ KEQEIHNNGI CLRTYNSTSF MEQRDHVLED RLLPQTELKP 300 FTDMKLGNDA ERRMFFADRS TSDLSMSVGL QGEKGHGSFS EGKFNGRNED QEGFKLQTVN 360 RLPYFETKLD LNVHEENDAA SSCKQFDLNG LSWN |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 2e-21 | 34 | 90 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 2e-21 | 34 | 90 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 2e-21 | 34 | 90 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 2e-21 | 34 | 90 | 1 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that may activate the transcription of specific genes involved in nitrogen uptake or assimilation (PubMed:15592750). Acts redundantly with MYR1 as a repressor of flowering and organ elongation under decreased light intensity (PubMed:21255164). Represses gibberellic acid (GA)-dependent responses and affects levels of bioactive GA (PubMed:21255164). {ECO:0000269|PubMed:21255164, ECO:0000305|PubMed:15592750}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00331 | DAP | Transfer from AT3G04030 | Download |
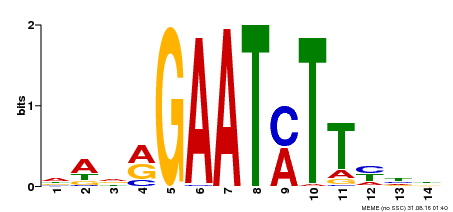
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Up-regulated by nitrogen deficiency. {ECO:0000269|PubMed:15592750}. | |||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017629911.1 | 0.0 | PREDICTED: myb-related protein 2-like | ||||
| Refseq | XP_017629912.1 | 0.0 | PREDICTED: myb-related protein 2-like | ||||
| Swissprot | Q9SQQ9 | 1e-149 | PHL9_ARATH; Myb-related protein 2 | ||||
| TrEMBL | A0A2P5YPP1 | 0.0 | A0A2P5YPP1_GOSBA; Uncharacterized protein | ||||
| STRING | Gorai.007G287500.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3336 | 27 | 64 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G04030.3 | 1e-148 | G2-like family protein | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



