 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cotton_A_23215_BGI-A2_v1.0 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 312aa MW: 35077.6 Da PI: 7.3709 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 105.9 | 2.1e-33 | 158 | 215 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-STT---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsagCpvkkkversaedpkvveitYegeHnhe 59
+Dgy+WrKYGqK vk+s+fprsYYrCt+ +C+vkk+ver+ +dp++v++tYeg+H+h+
Cotton_A_23215_BGI-A2_v1.0 158 EDGYRWRKYGQKAVKNSPFPRSYYRCTTISCNVKKRVERCFSDPSIVVTTYEGQHTHP 215
8********************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF038130 | 7.5E-110 | 1 | 312 | IPR017396 | PWRKY transcription factor, group IIc |
| Gene3D | G3DSA:2.20.25.80 | 9.0E-35 | 142 | 217 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 1.57E-29 | 149 | 217 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 31.238 | 152 | 217 | IPR003657 | WRKY domain |
| SMART | SM00774 | 1.2E-39 | 157 | 216 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 5.9E-26 | 158 | 215 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0009624 | Biological Process | response to nematode | ||||
| GO:0009733 | Biological Process | response to auxin | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 312 aa Download sequence Send to blast |
MERKEGVKVE DVLGNSSCSV NDFPLQSIFD LSSNEEEKIR SLGFMDLLGV QDLICSPVLE 60 IMAAQQVPSL MATQPPNPFS CTKIESPHEV FNQPATPNSS TISSASSEAA HDEPAKLDDQ 120 EEDQRKTKKQ LKPKKTNQKR QREPRFAFMT KSEVDHLEDG YRWRKYGQKA VKNSPFPRSY 180 YRCTTISCNV KKRVERCFSD PSIVVTTYEG QHTHPSPVMP RPNLVGSHLN SAISAASFGM 240 PMQTTPSHYH QHFQQPFTDN LSPLNFGHNG SLNATFLHQK RFCTPGPEPG PSLLKDHGLL 300 QDILPSHMLK EE |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj2_A | 6e-27 | 150 | 217 | 10 | 77 | Probable WRKY transcription factor 4 |
| 2lex_A | 6e-27 | 150 | 217 | 10 | 77 | Probable WRKY transcription factor 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor. Interacts specifically with the W box (5'-(T)TGAC[CT]-3'), a frequently occurring elicitor-responsive cis-acting element (By similarity). {ECO:0000250}. | |||||
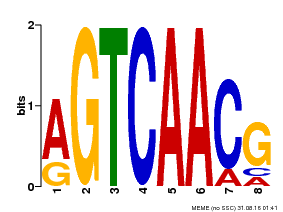
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00069 | PBM | Transfer from AT2G47260 | Download |
| |||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | KJ551845 | 0.0 | KJ551845.1 Gossypium hirsutum WRKY transcription factor 68 (WRKY68) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017629418.1 | 0.0 | PREDICTED: probable WRKY transcription factor 23 | ||||
| Swissprot | O22900 | 1e-69 | WRK23_ARATH; WRKY transcription factor 23 | ||||
| TrEMBL | A0A1U8L204 | 0.0 | A0A1U8L204_GOSHI; WRKY transcription factor | ||||
| STRING | Gorai.007G121200.1 | 0.0 | (Gossypium raimondii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6152 | 26 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G47260.1 | 2e-60 | WRKY DNA-binding protein 23 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



