 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cotton_A_36026_BGI-A2_v1.0 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Malvales; Malvaceae; Malvoideae; Gossypium
|
||||||||
| Family | ZF-HD | ||||||||
| Protein Properties | Length: 157aa MW: 18334 Da PI: 10.3297 | ||||||||
| Description | ZF-HD family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | ZF-HD_dimer | 65.9 | 7.5e-21 | 3 | 52 | 3 | 57 |
ZF-HD_dimer 3 kvrYkeClkNhAaslGghavDGCgEfmpsegeegtaaalkCaACgCHRnFHRrev 57
k Y++C++NhA lGg+a+DGC f+p + + ++C ACgCHRn+HR+
Cotton_A_36026_BGI-A2_v1.0 3 KEIYRDCQRNHARCLGGYAIDGCTQFIPF-----NNSKTHCKACGCHRNYHRKVL 52
678*************************9.....345689************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| ProDom | PD125774 | 1.0E-9 | 5 | 50 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| Pfam | PF04770 | 2.5E-16 | 5 | 51 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| TIGRFAMs | TIGR01566 | 1.3E-15 | 6 | 50 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| PROSITE profile | PS51523 | 20.916 | 6 | 51 | IPR006456 | ZF-HD homeobox protein, Cys/His-rich dimerisation domain |
| SuperFamily | SSF46689 | 1.43E-6 | 84 | 143 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 4.3E-14 | 85 | 144 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01565 | 3.0E-22 | 87 | 144 | IPR006455 | Homeodomain, ZF-HD class |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 157 aa Download sequence Send to blast |
MEKEIYRDCQ RNHARCLGGY AIDGCTQFIP FNNSKTHCKA CGCHRNYHRK VLCRDLEQSP 60 NGKSFRSLRE AKRMARQQQS CCGVNMKQRK SKFSMGQREA MREFGEGLGW SMRNKDRPEE 120 INRFCERIGV SRIIFKTWLN NNKKCYFKGT SSANISS |
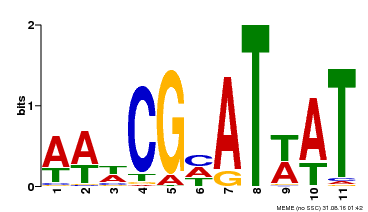
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00143 | DAP | Transfer from AT1G14687 | Download |
| |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_017633877.1 | 1e-114 | PREDICTED: zinc-finger homeodomain protein 10-like | ||||
| TrEMBL | A0A1U8MA23 | 1e-112 | A0A1U8MA23_GOSHI; zinc-finger homeodomain protein 10-like | ||||
| TrEMBL | A0A2P5WKH2 | 1e-112 | A0A2P5WKH2_GOSBA; Uncharacterized protein | ||||
| STRING | EOY01272 | 2e-65 | (Theobroma cacao) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM13728 | 21 | 24 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G14687.1 | 7e-35 | homeobox protein 32 | ||||



