 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cre03.g197350.t1.2 | ||||||||
| Common Name | CHLREDRAFT_129649 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Chlamydomonadales; Chlamydomonadaceae; Chlamydomonas
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 834aa MW: 91689.2 Da PI: 9.2115 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 40.7 | 5.5e-13 | 8 | 53 | 2 | 48 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
g W Ede+l+ av ++G ++W++I++ + ++++kqck rw+ +l
Cre03.g197350.t1.2 8 GVWKNTEDEILKAAVMKYGLNQWARISSLLV-RKSAKQCKARWYEWL 53
78*****************************.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 30.7 | 7.4e-10 | 60 | 102 | 2 | 47 |
SSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 2 grWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
WT eEde+l+++ k+++ W+tIa +g Rt+ qc +r+ +
Cre03.g197350.t1.2 60 TEWTREEDEKLLHLAKLMPCQ-WRTIAPIVG--RTPAQCLDRYERL 102
68*****************88.********8..*********9876 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 20.564 | 2 | 57 | IPR017930 | Myb domain |
| SMART | SM00717 | 2.1E-14 | 6 | 55 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 4.9E-18 | 9 | 60 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 4.33E-12 | 10 | 53 | No hit | No description |
| Pfam | PF13921 | 1.3E-12 | 10 | 70 | No hit | No description |
| SuperFamily | SSF46689 | 8.99E-20 | 33 | 107 | IPR009057 | Homeodomain-like |
| CDD | cd11659 | 4.07E-31 | 55 | 107 | No hit | No description |
| SMART | SM00717 | 8.6E-12 | 58 | 105 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS51294 | 15.747 | 58 | 107 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.1E-13 | 61 | 105 | IPR009057 | Homeodomain-like |
| Pfam | PF11831 | 9.6E-44 | 441 | 676 | IPR021786 | Pre-mRNA splicing factor component Cdc5p/Cef1 |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009870 | Biological Process | defense response signaling pathway, resistance gene-dependent | ||||
| GO:0010204 | Biological Process | defense response signaling pathway, resistance gene-independent | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0050832 | Biological Process | defense response to fungus | ||||
| GO:0009507 | Cellular Component | chloroplast | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 834 aa Download sequence Send to blast |
MRIMIKGGVW KNTEDEILKA AVMKYGLNQW ARISSLLVRK SAKQCKARWY EWLDPAIKKT 60 EWTREEDEKL LHLAKLMPCQ WRTIAPIVGR TPAQCLDRYE RLLDQAVAKD VNYDPRDDPR 120 RLRPGEIDPN PEAKPARPDA VDMDEDEKEM LSEARARLAN TRGKKAKRKA REKQLEEARR 180 LAQLQKKREL KAAGIDVRAR NKNSRAVDYS SEVAFELKPQ AGFYSTADEE KTTRSMQQEF 240 RPVTVEELEG RRRKDIEEAL IKKDVKRQKI NEMHDAPNAV AKHMGLDQEG PRRRGKLMLP 300 APQVSEAELE QIARLGVDGA LEASVSEGAG GDATRTLLGQ YNQTPAARLG MGPGATPMRT 360 PRAGPGGSDR IMAEAAALAR LQGMGTVLEG GEAGVDVAAM DFAGVTPRNV VAATPNPLAG 420 MTPSVRGAGG ATGARVVPSI AGVSATPSVA GTPLRAGSGG VGPGATPLIR DALGLNEADA 480 LTMEAAMSKR AAAARAAVMR GELRERLAGL PTPQNEYAFE VPELPEAEEE AEDAMEEDLA 540 DVRARKAREE AERRRIEELK KSKALQRQLP RPLNLDSLPA PKPAAPSTAG ASAEQLRDRA 600 EELVAAEMRG LLQHDAAKYP VKDGRDARKK GKQQHAAPGP VPVLEEFELE ALQSAAELVD 660 REVAYLRSAW DHAKLSPDDY SEVWMSVHRD LIYLPSRQRY ERAASATNVD RIDSLKSEFD 720 NVRADMEREA KKAAKLEGKL GLLLGGLQRR HGDLTGRVGE LWAQVRDAAQ ELVCFKALHE 780 RELRAAPERL EALGELVDAT KRREVDLQER FKALTRRRDE LAAALAQKRA AAS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5mqf_L | 0.0 | 2 | 825 | 3 | 802 | Cell division cycle 5-like protein |
| 5xjc_L | 0.0 | 2 | 825 | 3 | 802 | Cell division cycle 5-like protein |
| 5yzg_L | 0.0 | 2 | 825 | 3 | 802 | Cell division cycle 5-like protein |
| 5z56_L | 0.0 | 2 | 825 | 3 | 802 | Cell division cycle 5-like protein |
| 5z57_L | 0.0 | 2 | 825 | 3 | 802 | Cell division cycle 5-like protein |
| 5z58_L | 0.0 | 2 | 825 | 3 | 802 | Cell division cycle 5-like protein |
| 6ff4_L | 0.0 | 2 | 825 | 3 | 802 | Cell division cycle 5-like protein |
| 6ff7_L | 0.0 | 2 | 825 | 3 | 802 | Cell division cycle 5-like protein |
| 6icz_L | 0.0 | 2 | 825 | 3 | 802 | Cell division cycle 5-like protein |
| 6id0_L | 0.0 | 2 | 825 | 3 | 802 | Cell division cycle 5-like protein |
| 6id1_L | 0.0 | 2 | 825 | 3 | 802 | Cell division cycle 5-like protein |
| 6qdv_O | 0.0 | 2 | 825 | 3 | 802 | Cell division cycle 5-like protein |
| Search in ModeBase | ||||||
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Cre.2655 | 0.0 | normal conditions| nutrient deficiency | ||||
| Expression -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| Uniprot | TISSUE SPECIFICITY: Expressed extensively in shoot and root meristems. {ECO:0000269|PubMed:8917598}. | |||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
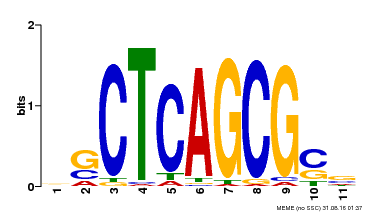
| UniProt | Component of the MAC complex that probably regulates defense responses through transcriptional control and thereby is essential for plant innate immunity. Possesses a sequence specific DNA sequence 'CTCAGCG' binding activity. Involved in mRNA splicing and cell cycle control. May also play a role in the response to DNA damage. {ECO:0000250|UniProtKB:Q99459, ECO:0000269|PubMed:17298883, ECO:0000269|PubMed:17575050, ECO:0000269|PubMed:8917598}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00028 | SELEX | Transfer from AT1G09770 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Cre03.g197350.t1.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_001693296.1 | 0.0 | predicted protein, partial | ||||
| Swissprot | P92948 | 0.0 | CDC5L_ARATH; Cell division cycle 5-like protein | ||||
| TrEMBL | A0A2K3DYR4 | 0.0 | A0A2K3DYR4_CHLRE; Uncharacterized protein | ||||
| STRING | EDP03322 | 0.0 | (Chlamydomonas reinhardtii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP1040 | 16 | 17 | Representative plant | OGRP3948 | 17 | 23 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G09770.1 | 0.0 | cell division cycle 5 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cre03.g197350.t1.2 |
| Entrez Gene | 5718778 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



