 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cre08.g361400.t1.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Chlamydomonadales; Chlamydomonadaceae; Chlamydomonas
|
||||||||
| Family | CPP | ||||||||
| Protein Properties | Length: 2022aa MW: 198776 Da PI: 7.8499 | ||||||||
| Description | CPP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | TCR | 43.9 | 4.9e-14 | 713 | 750 | 3 | 41 |
TCR 3 kkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNkee 41
k+C+Ckks+Clk+YC+Cfaag++C+ C+C +C+N+ e
Cre08.g361400.t1.2 713 AKSCRCKKSQCLKLYCDCFAAGQFCGA-CSCASCQNRPE 750
589***********************9.********876 PP
| |||||||
| 2 | TCR | 49.1 | 1.1e-15 | 784 | 824 | 1 | 41 |
TCR 1 kekkgCnCkkskClkkYCeCfaagkkCseeCkCedCkNkee 41
k+k+gCnCk+s+ClkkYCeCf+ g+kC+ +CkC +C+N ++
Cre08.g361400.t1.2 784 KHKRGCNCKRSHCLKKYCECFQGGVKCGMQCKCLECENMDD 824
589***********************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM01114 | 1.9E-11 | 711 | 751 | IPR033467 | Tesmin/TSO1-like CXC domain |
| PROSITE profile | PS51634 | 30.729 | 712 | 824 | IPR005172 | CRC domain |
| Pfam | PF03638 | 5.3E-11 | 714 | 748 | IPR005172 | CRC domain |
| SMART | SM01114 | 9.6E-15 | 784 | 825 | IPR033467 | Tesmin/TSO1-like CXC domain |
| Pfam | PF03638 | 8.9E-12 | 786 | 822 | IPR005172 | CRC domain |
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 2022 aa Download sequence Send to blast |
MRPESREQPK GQRRQATAHK PAEADLGSPL FLRNEVGNSP LPPMGSPLQY GVQSPCKYPL 60 FLESSTPRAT PRSAAPRDNN SKPSDLESSQ RMPFRSPFSP MRNHAPMALG ATQTPTPRRG 120 GVISVLTLLG GGRHTVAGAD IKPADGASHG AGGACAAAPS AAGTGSTTGA ASDGFAAASG 180 LGGGAGCGPG AGCQPGTEGP ALPPRSETPT AAGMAQCTAG FFSPNADMDA FFRLGSPMFA 240 TPARFTSTDL GSLAPFPLAG STPTAAMLLS PLPGLAGLGT LKSEVGADLA EPTGASGAGR 300 RRGRYLDFSG GAGAAGGEGG EVSGANAAAG QHQQHQQQQQ QQQDQQQMQL PARGLPPPLP 360 PLPPLPHFPS GSVNGGVPPP PGAPVLPPQG WGLPPPPALA MLQPPAPQQQ AGQPMQVDGA 420 EVRYSSQLPG MSSLLPFSSS TGGGSAGMGG VGVRGGLLST GSGSGSLDGS GGLGAGGGSG 480 NGGDMFGSRD SSPDMESGAG GAVGRGSAAR QVLLPPPAPL DVPPPSGVPS PVEYWDTVGV 540 DVRHPASGGD AVADAVANAL AAATAAASAV AVQHQHQSQR PSSSHQDASL PPAARQRNGG 600 GSTTTSASAP PPQPPPQQQP RQQPRSAAAA STAVAASPAT LQRPQRNRIA NTMLYGTTAV 660 VGGGSGGGGA GGRSTPSLMD ISVLDSDAPS SMDAKRSSAS GGNRARRPSD SGAKSCRCKK 720 SQCLKLYCDC FAAGQFCGAC SCASCQNRPE YADRVQQRRE DIAARDPQAF TRKIMDAPGG 780 GGGKHKRGCN CKRSHCLKKY CECFQGGVKC GMQCKCLECE NMDDDALGGG GGGGGAGGAG 840 AAAMAGAAKR TGNARQAAAA AAMSSGSWGR GGHAASVSAT ASDDDERSPP PPPDLPLPLP 900 LPAAAAMMAA AGGGGGLPPP PGLTSMASGG VVGGGGLSGY TDAGPEAGLP LPQPAAVEAM 960 VAAAAAAGAA SAVKPPRGGA PVGGGGSSKA GGGARKPPLP NSRGGRSRAK AKPWEMDTDD 1020 EEDAAAGMAA AEASDRGESG PEEGEDRLPL PLPASAGGAA GPAGLLPMPM PAPRHAQMQM 1080 PLGALPLPAQ QHQQPQQHAA AAAPPLPDSI LFPQRQAQAQ RHDTYQQQQQ QQHQQQQHQQ 1140 QQHNQQYQQQ HYHHQEQHAA DGQTARFGDL PGVKQEQEAA GLGATAAGHF SAPAQHHQHH 1200 QQHQQHQAPP QLEEAMQLPR IHTSFQPQSL SQHANPQQHH HHQQHQPHSA EPPVPSQHQP 1260 QQQQEQHQHP GPQQREFTQS GFQQQQPQQQ PQQQPRQPPT ATAAAAAAAL AADLSACTTP 1320 LSAALAAAGF NFDAGTPNAA AALAAFDLGD LPAALLMATP PTVGAGNAAA VAGGGGGGGV 1380 EREAAGADVK PGRDQRTADH TAADANADAH TVVRCQPAGS EPAAAGRGAG GAAAAACARS 1440 SLLPMSPMAP ATMLARHAIL AADSDDEDAG GDGGGGGRGG GAKVQGAAAA MAAAAAASAL 1500 AAGVVLPRQD QSQAAAAAAA EAGGGVAPMA ITPRRRGHGA SASLDLDFDV DLGLAMAEEE 1560 MAAAHPGDDF QALLGGGGGR NGGGGCGGAD ATLVCAELLE IHGSLDHMPP VPMPDSILGG 1620 GGALRVRTGA AADCSTPRRL SRFAPPGSAT AAAAAAAAAD SHFHGGLTAS RRAAAAAAAA 1680 AVATAGGSVH TLAAPVRTCS ASAAMAGVGA DARAHGASRI TGAKRKAGAA TGSDDDSDGD 1740 TEYRPDVPSP QHRSGNPARQ PSARQAARAA AASGAGAAKA GAKAGAKGGA AAAGGGGGTK 1800 SNGPDKEAMK AIALQYMTQR LGGGGPPCAP PGVPAPTPGP APAPGQLPLP AGLLPVPAPV 1860 PLSSVPGAGA AGIPPRPLAA AAQPPAGPPP PPPRAPSPAA VPAPDPPSHQ QHAAAEAGRR 1920 RPTSPDLCSP PATLTAPGAG LGLGLSQPPP HQQQQTQTAA STAPAPAAAA AAAGSSAGGG 1980 AAACGGGGGG KGLTCLTSLT RLTPMTKLGS GSTLFFSPGG R* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5fd3_A | 2e-25 | 705 | 824 | 7 | 123 | Protein lin-54 homolog |
| 5fd3_B | 2e-25 | 705 | 824 | 7 | 123 | Protein lin-54 homolog |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 1577 | 1584 | GGRNGGGG |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00624 | PBM | Transfer from PK22848.1 | Download |
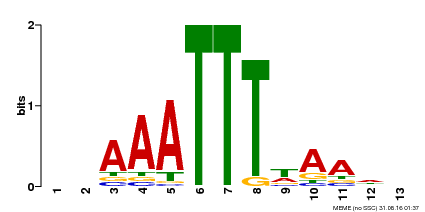
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Cre08.g361400.t1.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| TrEMBL | A0A2K3DGK7 | 0.0 | A0A2K3DGK7_CHLRE; Uncharacterized protein | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP323 | 15 | 30 | Representative plant | OGRP993 | 17 | 55 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G14770.1 | 4e-32 | TESMIN/TSO1-like CXC 2 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cre08.g361400.t1.2 |



