 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cre12.g501600.t1.2 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Chlorophyta; Chlorophyceae; Chlamydomonadales; Chlamydomonadaceae; Chlamydomonas
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 903aa MW: 89920.3 Da PI: 6.7069 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 27.9 | 5.1e-09 | 322 | 376 | 5 | 59 |
CHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 5 krerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaLkkeleelkkevakl 59
kr rr + NR +A +s +RK++++e+L + + e e+ L+++++++k +l
Cre12.g501600.t1.2 322 KRVRRIIANRMSAAKSKERKQQYTEQLSQMLDDTERERAGLQQQMDRYKVDNTRL 376
899********************************************99777665 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 1.4E-8 | 318 | 382 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF07716 | 1.5E-7 | 320 | 370 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.209 | 320 | 383 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 3.7E-9 | 320 | 380 | No hit | No description |
| SuperFamily | SSF57959 | 4.82E-8 | 322 | 378 | No hit | No description |
| CDD | cd14703 | 2.69E-16 | 323 | 374 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007231 | Biological Process | osmosensory signaling pathway | ||||
| GO:0008272 | Biological Process | sulfate transport | ||||
| GO:0009294 | Biological Process | DNA mediated transformation | ||||
| GO:0009652 | Biological Process | thigmotropism | ||||
| GO:0009970 | Biological Process | cellular response to sulfate starvation | ||||
| GO:0045596 | Biological Process | negative regulation of cell differentiation | ||||
| GO:0051170 | Biological Process | nuclear import | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005829 | Cellular Component | cytosol | ||||
| GO:0003682 | Molecular Function | chromatin binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0043621 | Molecular Function | protein self-association | ||||
| GO:0051019 | Molecular Function | mitogen-activated protein kinase binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 903 aa Download sequence Send to blast |
MSEWSQRLTV PAAAHRKTRS DTFGLMAMGA LNSWQEMPGL HTDAAPTSSP SGGEQGPDSS 60 SMDTEMAFVA VEADDAEMAG ATRERRKVSR RTGRAAGASS AAAGRQGSGA TATAAISAGL 120 VAPGISQGPE VGRAASSASL GSAGAWPLPH GPAPAGLQHP ACHTRSQSES FSDKLGGLAG 180 LQVGSFSSAR DAPASGMRQH VGAGGALLPL PPKAAASAQA RATSSGGASG SGGSLYNEAG 240 VGRSGMDEEL QDVEEEQELE DEDEELQDAA EEGGRRRGVK GGRSRSRGAR ASGGGDDSKG 300 GDKRGGGNNG RTQRELMLLD PKRVRRIIAN RMSAAKSKER KQQYTEQLSQ MLDDTERERA 360 GLQQQMDRYK VDNTRLEGYV EGVRREASSL TSTLTALRQQ GDELLRQLHA ARMAAAGGQS 420 PQAAGSAPGS EAPVPVPPEV DAAMKAAAAG AAAAAPPAAN FAELLSEGLT PEDLAAVERG 480 TLAVKAEPMA SQHQQAPARV PSATGMLVPR STTGVTQPPQ LVRPQHMAPG AAAPPQQAAL 540 SAQQQPQATN STSAAPPAAA TSAGGAPLHS MAPGVAPPGG FSHRPTQSAD AAAFMTFLPH 600 LGSLGPGNGA AVPGSSTLLP PGVGRGAFYA AAAASPPGAG PGSNDSSMLG LGLGVGAGGL 660 GPAAVAAMTS GPGSNSLSLP EPLDMTRARA AAEALAAAEA HGAAGGALGG LSVMAPGVAP 720 GAAPDMHTAA AMGMGLGMGM HGLGGAGTGM DAAVMSLSRL SAPMPGAQAL PVQLAGGAMQ 780 MPMGLVGSNS FGAGGGVGVG VPGALYAPGS AMAAAGVTVS AAAAAAVQQQ ALALQQQQLQ 840 LHQQQLHLQQ QQQMLALAAA GHLSAQQQQQ QQMDTTESFT IGRQVTTSLR PCVTTDPWRG 900 PA* |
| Expression -- UniGene ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| UniGene ID | E-value | Expressed in | ||||
| Cre.9229 | 0.0 | normal conditions| nutrient deficiency | ||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00129 | DAP | Transfer from AT1G06850 | Download |
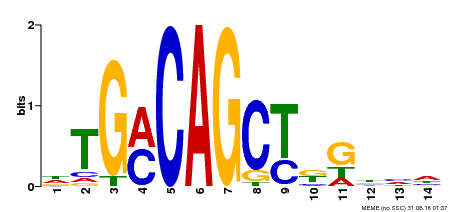
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Cre12.g501600.t1.2 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | - | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_001690784.1 | 1e-120 | predicted protein, partial | ||||
| TrEMBL | A0A2K3D369 | 0.0 | A0A2K3D369_CHLRE; Uncharacterized protein | ||||
| STRING | EDP05230 | 1e-119 | (Chlamydomonas reinhardtii) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Chlorophytae | OGCP6785 | 4 | 4 | Representative plant | OGRP126 | 17 | 181 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G06850.2 | 3e-10 | basic leucine-zipper 52 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cre12.g501600.t1.2 |



