 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Csa04g003010.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Camelina
|
||||||||
| Family | ARR-B | ||||||||
| Protein Properties | Length: 389aa MW: 43716.7 Da PI: 8.0717 | ||||||||
| Description | ARR-B family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 86.1 | 3.4e-27 | 198 | 251 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
k r++W+ eLHe+Fv+av+qL G++kA+Pk+ilelm+v+gL +e+v+SHLQk+Rl
Csa04g003010.1 198 KSRVVWSIELHEQFVNAVNQL-GIDKAVPKRILELMNVPGLNRENVASHLQKFRL 251
68*******************.********************************8 PP
| |||||||
| 2 | Response_reg | 74.5 | 4e-25 | 13 | 121 | 1 | 109 |
EEEESSSHHHHHHHHHHHHHTTCEEEEEESSHHHHHHHHHHHH..ESEEEEESSCTTSEHHHHHHHHHHHTTTSEEEEEESTTTHHHHHHHHHTT CS
Response_reg 1 vlivdDeplvrellrqalekegyeevaeaddgeealellkekd..pDlillDiempgmdGlellkeireeepklpiivvtahgeeedalealkaG 93
+l+vdD+ + + +l+++l + y +v+ +++++ al++l+e++ +Dl+l D+ mpgm+G++ll++ e +lp+i+++a g +++ ++ G
Csa04g003010.1 13 ILVVDDDTSCLFILEKMLLRLMY-QVTICSQADVALTILRERKgcFDLVLSDVHMPGMNGYKLLQQVGLEM-DLPVIMMSADGRTTTVMTGINHG 105
89*********************.*******************************************6644.8********************** PP
ESEEEESS--HHHHHH CS
Response_reg 94 akdflsKpfdpeelvk 109
a d+l Kp+ peel +
Csa04g003010.1 106 ACDYLIKPIRPEELKN 121
*************987 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PIRSF | PIRSF036392 | 1.9E-144 | 1 | 386 | IPR017053 | Response regulator B-type, plant |
| SuperFamily | SSF52172 | 2.13E-35 | 9 | 138 | IPR011006 | CheY-like superfamily |
| Gene3D | G3DSA:3.40.50.2300 | 5.8E-41 | 10 | 149 | No hit | No description |
| SMART | SM00448 | 1.3E-29 | 11 | 123 | IPR001789 | Signal transduction response regulator, receiver domain |
| PROSITE profile | PS50110 | 41.796 | 12 | 127 | IPR001789 | Signal transduction response regulator, receiver domain |
| Pfam | PF00072 | 4.3E-22 | 13 | 121 | IPR001789 | Signal transduction response regulator, receiver domain |
| CDD | cd00156 | 3.28E-27 | 14 | 126 | No hit | No description |
| PROSITE profile | PS51294 | 10.949 | 195 | 254 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 2.1E-28 | 195 | 256 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 1.47E-18 | 196 | 255 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.6E-23 | 198 | 251 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 1.6E-5 | 200 | 249 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000160 | Biological Process | phosphorelay signal transduction system | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0042802 | Molecular Function | identical protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 389 aa Download sequence Send to blast |
MAINDQFPNG LRILVVDDDT SCLFILEKML LRLMYQVTIC SQADVALTIL RERKGCFDLV 60 LSDVHMPGMN GYKLLQQVGL EMDLPVIMMS ADGRTTTVMT GINHGACDYL IKPIRPEELK 120 NIWQHVVRRK CVMNKEFRSS LAIEDNNKKN SGSVETVFSA SECSEGSLMK RRTKKKKRSV 180 DREDNENDDL LDPGNSKKSR VVWSIELHEQ FVNAVNQLGI DKAVPKRILE LMNVPGLNRE 240 NVASHLQKFR LYLKRLSGAA SQSKDTECIK RYENIQALVS SGQVHPQTLA ALFGQPIDNH 300 LSASFGVWIP NDDLGRSQTQ NFAVDVSSAS NRSVSVTNPM AVHDLSSSAN FSQKDDVNNN 360 TDHRIRQGYG SNVNEESWIL ERSSRQRY* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 2e-19 | 197 | 257 | 4 | 64 | ARR10-B |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 170 | 176 | RRTKKKK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-[AG]GATT-3'. Functions as a response regulator involved in His-to-Asp phosphorelay signal transduction system. Phosphorylation of the Asp residue in the receiver domain activates the ability of the protein to promote the transcription of target genes. Could directly activate some type-A response regulators in response to cytokinins (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00098 | PBM | Transfer from AT2G01760 | Download |
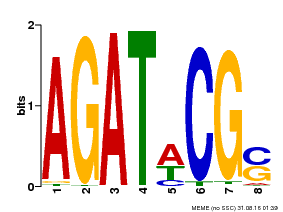
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Csa04g003010.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY099726 | 0.0 | AY099726.1 Arabidopsis thaliana putative two-component response regulator protein (At2g01760) mRNA, complete cds. | |||
| GenBank | AY128887 | 0.0 | AY128887.1 Arabidopsis thaliana putative two-component response regulator protein (At2g01760) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010502166.1 | 0.0 | PREDICTED: two-component response regulator ARR14-like isoform X2 | ||||
| Swissprot | Q8L9Y3 | 0.0 | ARR14_ARATH; Two-component response regulator ARR14 | ||||
| TrEMBL | R0H786 | 0.0 | R0H786_9BRAS; Two-component response regulator | ||||
| STRING | XP_010502166.1 | 0.0 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM8150 | 27 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G01760.1 | 0.0 | response regulator 14 | ||||



