 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Csa04g031200.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Camelina
|
||||||||
| Family | E2F/DP | ||||||||
| Protein Properties | Length: 410aa MW: 46317.8 Da PI: 7.4348 | ||||||||
| Description | E2F/DP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | E2F_TDP | 72.3 | 5.3e-23 | 36 | 101 | 1 | 71 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvsedvknkrRRiYDilNVLealnliekkekneirwkg 71
rk+ksL+ll+++fl+l+++ +++ l+++a +L v +rRRiYDi+NVLe+++++++++kn+++wkg
Csa04g031200.1 36 RKQKSLGLLCTNFLALYNRDGIDTIGLDDAAFKL---GV--ERRRIYDIVNVLESVGVLTRRAKNQYTWKG 101
79*****************999************...**..****************************98 PP
| |||||||
| 2 | E2F_TDP | 79.9 | 2.2e-25 | 177 | 257 | 1 | 71 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvse.....dvknkrRRiYDilNVLealnliek.....kekneirwkg 71
r+eksL+lltq+f+kl+ +se++i++l+e+ak L+ + +++k+RR+YDi+NVL+++nliek ++k++++w g
Csa04g031200.1 177 RREKSLGLLTQNFIKLFICSEARIISLDEAAKLLLGDahntsIMRTKVRRLYDIANVLSSMNLIEKthtldSRKPAFKWLG 257
689**************************************999************************9999*******87 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 4.4E-25 | 32 | 102 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 4.46E-16 | 35 | 99 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 4.4E-27 | 36 | 101 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Pfam | PF02319 | 4.2E-20 | 37 | 101 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Gene3D | G3DSA:1.10.10.10 | 2.1E-23 | 175 | 258 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 1.7E-32 | 177 | 257 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| SuperFamily | SSF46785 | 5.9E-14 | 179 | 258 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| Pfam | PF02319 | 5.4E-20 | 179 | 257 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032876 | Biological Process | negative regulation of DNA endoreduplication | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005667 | Cellular Component | transcription factor complex | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 410 aa Download sequence Send to blast |
MSDLPPERSI VPITSSSPVY MPESSSYLQL HHSYSRKQKS LGLLCTNFLA LYNRDGIDTI 60 GLDDAAFKLG VERRRIYDIV NVLESVGVLT RRAKNQYTWK GFAAIPAALK ELQEVFLMQE 120 EAVKDTFHRF YVNENVKGSD DEDDDEESSQ PHSSSQTDSS KPGSLPQSSD SPKLDNRREK 180 SLGLLTQNFI KLFICSEARI ISLDEAAKLL LGDAHNTSIM RTKVRRLYDI ANVLSSMNLI 240 EKTHTLDSRK PAFKWLGYNG EPTFTLSSDL LQMESRKRAF GTDLTNVNVK RSKSSSSSQD 300 NATERRLKMK KHSTAESSYN KSFDVYESRH GSTGYQFGPF APGTGTYPTA VLEDNSRRAF 360 DVENLVSDHH PSYQNQVLKD LFSHYMDAWN TWYSEVIQKN PLPNASQHR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4yo2_A | 8e-38 | 35 | 249 | 3 | 228 | Transcription factor E2F8 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Inhibitor of E2F-dependent activation of gene expression. Binds specifically the E2 recognition site without interacting with DP proteins and prevents transcription activation by E2F/DP heterodimers. Controls the timing of endocycle onset and inhibits endoreduplication. {ECO:0000269|PubMed:11786543, ECO:0000269|PubMed:11867638, ECO:0000269|PubMed:15649366, ECO:0000269|PubMed:18787127}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00397 | DAP | Transfer from AT3G48160 | Download |
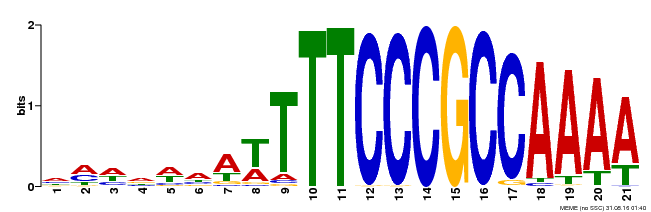
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Csa04g031200.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AB074533 | 0.0 | AB074533.1 Arabidopsis thaliana mRNA for E2F-like repressor E2L3, complete cds. | |||
| GenBank | BT004258 | 0.0 | BT004258.1 Arabidopsis thaliana clone RAFL15-19-A14 (R20743) putative DP-E2F protein 1 (At3g48160) mRNA, complete cds. | |||
| GenBank | BT005524 | 0.0 | BT005524.1 Arabidopsis thaliana clone U20743 putative DP-E2F protein 1 (At3g48160) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010503439.1 | 0.0 | PREDICTED: E2F transcription factor-like E2FE | ||||
| Swissprot | Q8LSZ4 | 0.0 | E2FE_ARATH; E2F transcription factor-like E2FE | ||||
| TrEMBL | D7LRJ4 | 0.0 | D7LRJ4_ARALL; Uncharacterized protein | ||||
| STRING | XP_010503439.1 | 0.0 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM6304 | 27 | 43 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G48160.2 | 0.0 | DP-E2F-like 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



