 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Csa06g044610.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Camelina
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 412aa MW: 46342.8 Da PI: 6.84 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 100.7 | 9.7e-32 | 86 | 139 | 2 | 55 |
G2-like 2 prlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
prlrWtpeLH +F++ave+LGG+++AtPk +l+lm+vkgL+++hvkSHLQ+YR+
Csa06g044610.1 86 PRLRWTPELHICFLQAVERLGGPDRATPKLVLQLMNVKGLSIAHVKSHLQMYRS 139
8****************************************************7 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 13.117 | 82 | 142 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.8E-30 | 82 | 140 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 5.02E-16 | 83 | 141 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 2.9E-24 | 86 | 141 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 2.3E-9 | 87 | 138 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 412 aa Download sequence Send to blast |
MRSSSKSSEN SKTCLSNNFK ATTNIEEDKD EEEDEEGEED EEERSGDQSP SSNSYVEESG 60 SHQNHDHDQI KKNGGSVRPY NRSKTPRLRW TPELHICFLQ AVERLGGPDR ATPKLVLQLM 120 NVKGLSIAHV KSHLQMYRSK KTDDPNQGDQ GFSFEHGAGY TYNLSQLPML QSFDQRPSSS 180 LGYGSGSWTD HRRQIYRSPW RGLTARDTTR TRQTLFSPRP GERVHGVSNS ILNDKNKTIS 240 FRINSHEAAN ENNGVAEVVP RIHRSFLEGM KTFNKSWGQS LSSNPNPSTA TRPQDHSATT 300 RNSYQWENAR VAEGSENVLK RKRVLLSDDC NTSNQDLDLS LSLKVPRTHD NLGECLLENE 360 EKEHDNHHDS LQGLSLSLSS SGLSKLGRTI RKEDQTERKI EVLASPLDLT L* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4r_A | 3e-17 | 87 | 141 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 3e-17 | 87 | 141 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 3e-17 | 87 | 141 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 3e-17 | 87 | 141 | 3 | 57 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00301 | DAP | Transfer from AT2G40260 | Download |
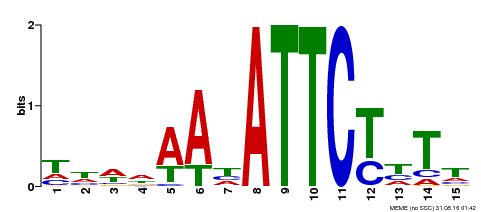
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Csa06g044610.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AC254595 | 0.0 | AC254595.1 Capsella rubella clone CAP16-M18, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010517442.1 | 0.0 | PREDICTED: uncharacterized protein LOC104792882 | ||||
| TrEMBL | R0FW67 | 0.0 | R0FW67_9BRAS; Uncharacterized protein | ||||
| STRING | XP_010517442.1 | 0.0 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM9359 | 21 | 36 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G40260.1 | 0.0 | G2-like family protein | ||||



