 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Csa08g002660.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Camelina
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 956aa MW: 106720 Da PI: 5.0948 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 56.3 | 7.3e-18 | 34 | 80 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg WT+eEde l +av+ + g++Wk+Ia+++ Rt+ qc +rwqk+l
Csa08g002660.1 34 RGQWTAEEDEVLRKAVHSFKGKNWKKIAEYFK-DRTDVQCLHRWQKVL 80
89*****************************9.************986 PP
| |||||||
| 2 | Myb_DNA-binding | 64.3 | 2.4e-20 | 86 | 132 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
+g+WT+eEde +v++++++G++ W+tIar+++ gR +kqc++rw+++l
Csa08g002660.1 86 KGPWTKEEDEMIVQLIEKYGPKKWSTIARYLP-GRIGKQCRERWHNHL 132
79******************************.*************97 PP
| |||||||
| 3 | Myb_DNA-binding | 42.6 | 1.4e-13 | 138 | 180 | 1 | 45 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHH CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwq 45
+ +WT+eE++ l++a++ +G++ W+ + ++ gR+++ +k++w+
Csa08g002660.1 138 KEAWTQEEELVLIRAHQIYGNK-WAELTKFLP-GRSDNGIKNHWH 180
579*******************.*********.***********8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 18.899 | 29 | 80 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 2.8E-15 | 31 | 86 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 8.7E-15 | 33 | 82 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.2E-15 | 34 | 80 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-22 | 35 | 88 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 1.67E-13 | 37 | 80 | No hit | No description |
| PROSITE profile | PS51294 | 33.057 | 81 | 136 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 1.39E-31 | 83 | 179 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 1.5E-19 | 85 | 134 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 3.1E-18 | 86 | 132 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.62E-17 | 88 | 132 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.5E-27 | 89 | 136 | IPR009057 | Homeodomain-like |
| Gene3D | G3DSA:1.10.10.60 | 1.3E-19 | 137 | 187 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 18.044 | 137 | 187 | IPR017930 | Myb domain |
| SMART | SM00717 | 4.4E-12 | 137 | 185 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 1.1E-11 | 138 | 180 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.12E-9 | 140 | 180 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0032875 | Biological Process | regulation of DNA endoreduplication | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003713 | Molecular Function | transcription coactivator activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 956 aa Download sequence Send to blast |
MESESTTTTI STPQDGIPKS RHGRTSGPAR RSTRGQWTAE EDEVLRKAVH SFKGKNWKKI 60 AEYFKDRTDV QCLHRWQKVL NPELVKGPWT KEEDEMIVQL IEKYGPKKWS TIARYLPGRI 120 GKQCRERWHN HLNPAINKEA WTQEEELVLI RAHQIYGNKW AELTKFLPGR SDNGIKNHWH 180 SSVKKKLDSY MSSGLLDQYQ AMPLAPYERS STLQSAFMQS NIDGNGEIDS RQNSSMVGCS 240 LSAGDFSNGA INMDHHFYPC RNSQETEQAV YHSDQYFYPE LEDISVSISE VSYDMEDCSQ 300 FPDHNNVSTA PSQDYQFDFQ ELSDISLEMR QANMSELPMP YTKESKESTV EPTNSTLSID 360 VATYTNSANV LTPETECCRV LFPDQESEGH SVSRSLTQEP NEVNQSDSRD PILSASASDS 420 QISEATKSPM QSSSSRLNAT AASGKETLRP APLIITPDKY AKKSSGLICH PFEAEPKCTT 480 NGNGRFICID DPSSFTCVDG GTNNSSEEDQ SYHVNDSKKL VAVSDFASLA EDRPHSPPKH 540 DPNTTSEQHH EDMGASSSLG FPSLDLPVFS CDLLQSKNDP LHDYSPLGIR KLLMSTMTCI 600 SPIRLWGSPT GKKTLVGAKS ILRKRTRDLL TPLSEKRSDK KLETDIAASL AKDFSRLDVM 660 FDESENRESI FGNNTDVILG DRINSGEGEE NWRSNPSSLF SHKMPEETVH IRKSLEKAEQ 720 TCMEANVREN DGSGANVENA ESFSGVLSEL NNCKPVLSSP PGQSVTKAEK AQVLTPRNQF 780 QKTFMANKEQ HSSSSLCLVI NSPSLARNTE GHLVDNETSN ENFSIFCGTP FRRGLESPSA 840 WKSPFYVNSL LPSPRFDTEI TIEDMGYIFS PGERSYDSIG VMTQRNEHAR AFAAFADAME 900 VSISPTNDDA EQKKELNKEN NDPLLAERRV LDFNDCESPV KATEEVSSYL LKGSR* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1h88_C | 3e-68 | 34 | 187 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| 1h89_C | 3e-68 | 34 | 187 | 6 | 159 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds 5'-AACGG-3' motifs in gene promoters (PubMed:21862669). Involved in the regulation of cytokinesis, probably via the activation of several G2/M phase-specific genes transcription (e.g. KNOLLE) (PubMed:17287251, PubMed:21862669, PubMed:25806785). Required for the maintenance of diploidy (PubMed:21862669). {ECO:0000269|PubMed:17287251, ECO:0000269|PubMed:21862669, ECO:0000269|PubMed:25806785}.; FUNCTION: Involved in transcription regulation during induced endoreduplication at the powdery mildew (e.g. G.orontii) infection site, thus promoting G.orontii growth and reproduction. {ECO:0000269|PubMed:20018666}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00505 | DAP | Transfer from AT5G11510 | Download |
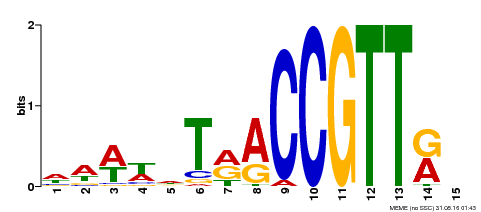
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Csa08g002660.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly induced by salicylic acid (SA) (PubMed:16463103). Expressed in a cell cycle-dependent manner, with highest levels 2 hours before the peak of mitotic index in cells synchronized by aphidicolin. Activated by CYCB1 (PubMed:17287251). Accumulates at powdery mildew (e.g. G.orontii) infected cells. {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:17287251}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AF371975 | 0.0 | AF371975.2 Arabidopsis thaliana putative c-myb-like transcription factor MYB3R-4 (MYB3R4) mRNA, complete cds. | |||
| GenBank | AY519650 | 0.0 | AY519650.1 Arabidopsis thaliana MYB transcription factor (At5g11510) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019083774.1 | 0.0 | PREDICTED: myb-related protein 3R-1-like | ||||
| Swissprot | Q94FL9 | 0.0 | MB3R4_ARATH; Transcription factor MYB3R-4 | ||||
| TrEMBL | R0H5A0 | 0.0 | R0H5A0_9BRAS; Uncharacterized protein | ||||
| STRING | XP_010419723.1 | 0.0 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM13213 | 18 | 28 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G11510.1 | 0.0 | myb domain protein 3r-4 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



