 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Csa08g055230.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Camelina
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1021aa MW: 114756 Da PI: 5.7513 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 184.7 | 9.8e-58 | 12 | 129 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfq 95
++e ++rwl++ ei++iL+n++k+++++e++ rp sgsl+L++rk++ryfrkDG++w+kkkdgkt++E+hekLKvg+v+vl+cyYah+e n++fq
Csa08g055230.1 12 MSEaRHRWLRPAEICEILQNHHKFHIASESPPRPASGSLFLFDRKVLRYFRKDGHNWRKKKDGKTIKEAHEKLKVGSVDVLHCYYAHGEGNENFQ 106
45559****************************************************************************************** PP
CG-1 96 rrcywlLeeelekivlvhylevk 118
rrcyw+Le++l +iv+vhylevk
Csa08g055230.1 107 RRCYWMLEQDLMHIVFVHYLEVK 129
********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 84.651 | 8 | 134 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 1.8E-84 | 11 | 129 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 8.6E-51 | 14 | 128 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 2.64E-14 | 425 | 511 | IPR014756 | Immunoglobulin E-set |
| Pfam | PF01833 | 2.1E-4 | 425 | 500 | IPR002909 | IPT domain |
| Gene3D | G3DSA:2.60.40.10 | 5.0E-4 | 426 | 500 | IPR013783 | Immunoglobulin-like fold |
| Pfam | PF12796 | 2.2E-7 | 609 | 686 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 19.227 | 609 | 720 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 6.4E-18 | 610 | 721 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 4.04E-19 | 610 | 720 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 5.49E-14 | 615 | 718 | No hit | No description |
| PROSITE profile | PS50088 | 11.22 | 659 | 691 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.0029 | 659 | 688 | IPR002110 | Ankyrin repeat |
| SuperFamily | SSF52540 | 9.56E-8 | 833 | 884 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| SMART | SM00015 | 0.51 | 834 | 856 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.657 | 835 | 864 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.0024 | 837 | 855 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.047 | 857 | 879 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.359 | 858 | 882 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 0.003 | 860 | 879 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0050826 | Biological Process | response to freezing | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005516 | Molecular Function | calmodulin binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1021 aa Download sequence Send to blast |
MHITRIDMEQ LMSEARHRWL RPAEICEILQ NHHKFHIASE SPPRPASGSL FLFDRKVLRY 60 FRKDGHNWRK KKDGKTIKEA HEKLKVGSVD VLHCYYAHGE GNENFQRRCY WMLEQDLMHI 120 VFVHYLEVKG NRTSQGMKEN NSNSVNGTSS VNIDSTASPT STLSSLCEDA DSGDSHQARS 180 VLRASPESQT GNCYGWTPPT SMHNVSQVHG NRVRESDSQR SVDVRAWDAV GNSLTRYHDP 240 PYCNNLLTQM QPSITDSMLV DENTDKGGRL KSEPIRNPLQ MQSNWQIPVQ DDLLLPKGQE 300 DLFSHSGMTD DIDLALFELS AEDNFETFAS LLGSENQQPF GNSYQAPPSS MESEYIPVKK 360 SLLRSEDSLK KVDSFSRWAI KELAEMEDLQ MQSSREDIAW TTVECETAAA GISLSPSLSE 420 DQRFTIVDFW PKCAETDAEV EVMVIGTFLL SPQEVTKYNW SCMFGEVEVP AEILVDGVLC 480 CHAPPHTAGH VPFYVTCSNR FACSEVREFD FLSGSTQKID AADVHGPSTD EAASLQLRFE 540 KLLAHRAFVN EYHIFEDVGE KRGKISKIML LKEEKEYLLP GTYQRDSTKQ EPKGQLFREQ 600 LEEKLYIWLI HKVSEEGKGP NMLDEDGQGI LHFIAALGYD WAIKPMLAAG VNINFRDANG 660 WSALHWAAFS GREETVAVLV SLGADAGALT DPSPELPLGK TAADLAYANG HRGISGFLAE 720 SSLTSYLEKL TVDSKENSSA NSFGEKAVQT VSERSAAPMS YGDLPEKLSL KDSLTAVRNA 780 TQAADRLHQV FRMQSFERKQ LSEFVDDDKI DISDELAVSF AASKTKTPGH SDVSLSSAAT 840 HIQKKYRGWK KRKEFLLIRQ RIVRIQAHVR GHQVRKQYRT VIWSVGLLEK IILRWRRKGK 900 GLRGFKSSAI AKPVEPEPTA SAVSPTIPQE DEYDFLKEGR KQTEERLQKA LTRVKSMVQY 960 PEARDQYRRL LTVVEGFREN EASSSASINN EVGPVNCDED DLIDIDSLLN DDTLMMSFSP 1020 * |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
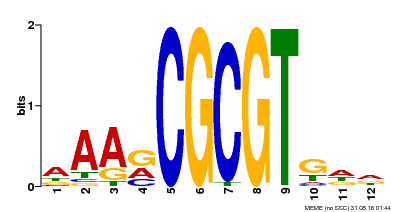
| UniProt | Transcription activator that binds calmodulin in a calcium-dependent manner in vitro (PubMed:11925432). Binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Involved in freezing tolerance (PubMed:19270186). Involved in freezing tolerance in association with CAMTA2 and CAMTA3. Contributes together with CAMTA2 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved in drought stress responses by regulating several drought-responsive genes (PubMed:23547968). Involved in auxin signaling and responses to abiotic stresses (PubMed:20383645). Activates the expression of the V-PPase proton pump AVP1 in pollen (PubMed:14581622). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:11925432, ECO:0000269|PubMed:14581622, ECO:0000269|PubMed:19270186, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:23547968, ECO:0000269|PubMed:23581962, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00501 | DAP | Transfer from AT5G09410 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Csa08g055230.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by UVB, wounding, ethylene and methyl jasmonate (PubMed:12218065). Induced by salt stress and heat shock (PubMed:12218065, PubMed:20383645). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:20383645, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK228740 | 0.0 | AK228740.1 Arabidopsis thaliana mRNA for Calmodulin-binding transcription activator 1, complete cds, clone: RAFL16-07-H14. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019084442.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 1 isoform X2 | ||||
| Swissprot | Q9FY74 | 0.0 | CMTA1_ARATH; Calmodulin-binding transcription activator 1 | ||||
| TrEMBL | R0FCP0 | 0.0 | R0FCP0_9BRAS; Uncharacterized protein | ||||
| STRING | XP_010422986.1 | 0.0 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4082 | 24 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G09410.2 | 0.0 | ethylene induced calmodulin binding protein | ||||



