 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Csa09g082590.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Camelina
|
||||||||
| Family | Trihelix | ||||||||
| Protein Properties | Length: 575aa MW: 65870.3 Da PI: 7.8047 | ||||||||
| Description | Trihelix family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | trihelix | 97.2 | 1.5e-30 | 37 | 121 | 1 | 87 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqlea 87
rW++ e+laL+++r+em++++r++ lk+plWee+s+km+e g++rs+k+Ckek+en+ k++k++k+g+ ++++++s +++f++lea
Csa09g082590.1 37 RWPRPETLALLRIRSEMDKAFRDSTLKAPLWEEISRKMMELGYKRSAKKCKEKFENVYKYHKRTKDGRTGKSEGKS--YRFFEELEA 121
8********************************************************************9877776..******985 PP
| |||||||
| 2 | trihelix | 108.8 | 3.5e-34 | 396 | 481 | 1 | 87 |
trihelix 1 rWtkqevlaLiearremeerlrrgklkkplWeevskkmrergferspkqCkekwenlnkrykkikegekkrtsessstcpyfdqlea 87
rW+k ev aLi++r+++e +++++ k+plWee+s+ mr+ g++rs+k+Ckekwen+nk++kk+ke++kkr + +s+tcpyf+qlea
Csa09g082590.1 396 RWPKTEVEALIRIRKNLEANYQENGTKGPLWEEISAGMRRLGYNRSAKRCKEKWENINKYFKKVKESNKKR-PLDSKTCPYFHQLEA 481
8*********************************************************************8.9************85 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00717 | 0.044 | 34 | 96 | IPR001005 | SANT/Myb domain |
| PROSITE profile | PS50090 | 7.004 | 36 | 94 | IPR017877 | Myb-like domain |
| CDD | cd12203 | 4.26E-25 | 36 | 101 | No hit | No description |
| Pfam | PF13837 | 1.9E-20 | 36 | 121 | No hit | No description |
| PROSITE profile | PS50090 | 7.526 | 389 | 453 | IPR017877 | Myb-like domain |
| SMART | SM00717 | 9.2E-4 | 393 | 455 | IPR001005 | SANT/Myb domain |
| CDD | cd12203 | 3.85E-26 | 395 | 460 | No hit | No description |
| Gene3D | G3DSA:1.10.10.60 | 3.1E-4 | 395 | 452 | IPR009057 | Homeodomain-like |
| Pfam | PF13837 | 7.1E-24 | 395 | 482 | No hit | No description |
| SuperFamily | SSF46689 | 6.22E-5 | 396 | 487 | IPR009057 | Homeodomain-like |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 575 aa Download sequence Send to blast |
MSGNSSGPLE SSGGGVGGSG EEEKDMKMEE TGGGGNRWPR PETLALLRIR SEMDKAFRDS 60 TLKAPLWEEI SRKMMELGYK RSAKKCKEKF ENVYKYHKRT KDGRTGKSEG KSYRFFEELE 120 AFETLNSYQH EPDQSHPAKS SAAVATAAIT TSLIPWISSN NPSTEKSSLP LKHQHQVSVQ 180 PITTNPTFLA KQPSATMPFP FYSNNNTTTV SQPPPISNDL MNNVSSLHLF SSSTSSSTAS 240 DEEEDHHQGK RSRKRRKYWK GLFTKLTKEL MEKQEKMQKR FLETLENREK ERISREEAWR 300 VQEIARINRE HETLIHERSN AAAKDAAIIS FLHKISGGQQ QQPQQQNHHK PAQRKQYQSD 360 HSITFESKEP RPILLDTTMK MGNYDTNHSI SPSSSRWPKT EVEALIRIRK NLEANYQENG 420 TKGPLWEEIS AGMRRLGYNR SAKRCKEKWE NINKYFKKVK ESNKKRPLDS KTCPYFHQLE 480 ALYNERNKSG AMPLPLPLMV TPQRQLLLSQ ETQTEFETDQ RDKVGDKEDN EEGESEEDEY 540 DDEEEGEGDN ETSEFETVLK KTSSPMDINN NLFT* |
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 79 | 87 | KRSAKKCKE |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probable transcription factor that binds specific DNA sequence. {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00244 | DAP | Transfer from AT1G76890 | Download |
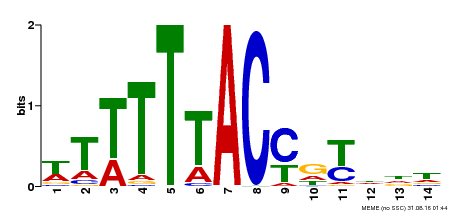
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Csa09g082590.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY087117 | 0.0 | AY087117.1 Arabidopsis thaliana clone 3190 mRNA, complete sequence. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010428754.1 | 0.0 | PREDICTED: trihelix transcription factor GT-2-like | ||||
| Swissprot | Q39117 | 0.0 | TGT2_ARATH; Trihelix transcription factor GT-2 | ||||
| TrEMBL | A0A178W7Z2 | 0.0 | A0A178W7Z2_ARATH; GT2 | ||||
| STRING | XP_010428754.1 | 0.0 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM5995 | 28 | 47 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G76890.2 | 0.0 | Trihelix family protein | ||||



