 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Csa11g065130.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Camelina
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 462aa MW: 51974.9 Da PI: 6.5275 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 38.5 | 2e-12 | 293 | 338 | 4 | 54 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksL 54
+h e+Er RR+++N++f Lr ++P+ K++K + Le Av+YI++L
Csa11g065130.1 293 NHVEAERMRREKLNHRFYALRAVVPNV-----SKMDKTSLLEDAVRYINEL 338
799***********************6.....5***************998 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 2.5E-50 | 35 | 227 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| CDD | cd00083 | 1.51E-13 | 288 | 339 | No hit | No description |
| SuperFamily | SSF47459 | 4.58E-17 | 289 | 360 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 16.806 | 289 | 338 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene3D | G3DSA:4.10.280.10 | 5.2E-16 | 293 | 360 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 7.2E-10 | 293 | 338 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 3.8E-14 | 295 | 344 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042538 | Biological Process | hyperosmotic salinity response | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0005509 | Molecular Function | calcium ion binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 462 aa Download sequence Send to blast |
MNTDDNLSMI EALLTSDLWP PLPPAKHHLS LETTLQKRLH AVLSGTHEPW TYAIFWKPSD 60 FDLSGESVLK WGDGVYNGDD EEKTGRRLRR TIKKTIPSSP EEKERWSKVL RELNSMISGE 120 PFPVVEEDDA KDDVEVTDTE WFFLVSMTWT FGNGSGLAGK AFTTYNPVWV TGSDQIYGSG 180 CDRAKQGGVL GLQTIVCIPS DNGVLELGST EQISQNSNLF NKIRYLFSFE GSKYFSGAPT 240 SAQFRSSIHN PNPNPVYQPI QINLSGEEMV IPEMKPGKKR GRKPMHGREK PLNHVEAERM 300 RREKLNHRFY ALRAVVPNVS KMDKTSLLED AVRYINELKS TAENAESERN AVQNQLNELK 360 KITGRQNATS SVVCNDDKEN VSELKIEVKA MGSDVMIRVE SGKRNHPGAR LMNALMDLEL 420 EVSHASISVM NDLMIQQATV KMGLRIYEQE QLRDMLKSKI N* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 4rqw_A | 7e-50 | 32 | 229 | 6 | 194 | Transcription factor MYC3 |
| 4rqw_B | 7e-50 | 32 | 229 | 6 | 194 | Transcription factor MYC3 |
| 4rs9_A | 7e-50 | 32 | 229 | 6 | 194 | Transcription factor MYC3 |
| 4yz6_A | 7e-50 | 32 | 229 | 6 | 194 | Transcription factor MYC3 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00547 | DAP | Transfer from AT5G46830 | Download |
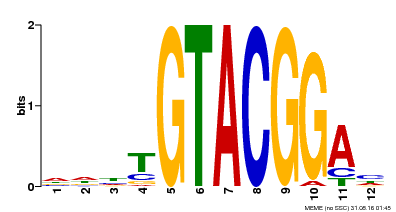
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Csa11g065130.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By UV treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010441529.1 | 0.0 | PREDICTED: transcription factor bHLH28-like | ||||
| Swissprot | Q9LUK7 | 0.0 | BH028_ARATH; Transcription factor bHLH28 | ||||
| TrEMBL | R0G888 | 0.0 | R0G888_9BRAS; Uncharacterized protein | ||||
| STRING | XP_010441529.1 | 0.0 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM13953 | 16 | 24 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G46830.1 | 0.0 | NACL-inducible gene 1 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



