 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Csa11g101340.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Camelina
|
||||||||
| Family | CAMTA | ||||||||
| Protein Properties | Length: 1038aa MW: 115567 Da PI: 6.1907 | ||||||||
| Description | CAMTA family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | CG-1 | 188.3 | 7.6e-59 | 19 | 136 | 2 | 118 |
CG-1 2 lke.kkrwlkneeiaaiLenfekheltlelktrpksgsliLynrkkvryfrkDGyswkkkkdgktvrEdhekLKvggvevlycyYahseenptfq 95
l+e ++rwl++ ei++iL n++k+++++e+++rp+sgsl+L++rk++ryfrkDG++w+kkkdgktv+E+hekLKvg+++vl+cyYah+e+n++fq
Csa11g101340.1 19 LSEaQHRWLRPAEICEILRNHQKFHIASEPPNRPPSGSLFLFDRKVLRYFRKDGHNWRKKKDGKTVKEAHEKLKVGSIDVLHCYYAHGEDNENFQ 113
45559****************************************************************************************** PP
CG-1 96 rrcywlLeeelekivlvhylevk 118
rrcyw+Le++l +iv+vhylevk
Csa11g101340.1 114 RRCYWMLEQDLMHIVFVHYLEVK 136
********************985 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51437 | 86.275 | 15 | 141 | IPR005559 | CG-1 DNA-binding domain |
| SMART | SM01076 | 3.6E-87 | 18 | 136 | IPR005559 | CG-1 DNA-binding domain |
| Pfam | PF03859 | 8.2E-52 | 21 | 135 | IPR005559 | CG-1 DNA-binding domain |
| SuperFamily | SSF81296 | 4.06E-13 | 446 | 530 | IPR014756 | Immunoglobulin E-set |
| SuperFamily | SSF48403 | 7.15E-19 | 625 | 738 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 19.412 | 627 | 738 | IPR020683 | Ankyrin repeat-containing domain |
| Pfam | PF12796 | 1.8E-7 | 628 | 704 | IPR020683 | Ankyrin repeat-containing domain |
| Gene3D | G3DSA:1.25.40.20 | 6.4E-18 | 628 | 739 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 1.86E-14 | 639 | 729 | No hit | No description |
| SMART | SM00248 | 1600 | 644 | 673 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 8.843 | 644 | 676 | IPR002110 | Ankyrin repeat |
| SMART | SM00248 | 0.0029 | 677 | 706 | IPR002110 | Ankyrin repeat |
| PROSITE profile | PS50088 | 10.873 | 677 | 709 | IPR002110 | Ankyrin repeat |
| SMART | SM00015 | 0.32 | 852 | 874 | IPR000048 | IQ motif, EF-hand binding site |
| SuperFamily | SSF52540 | 8.23E-8 | 853 | 902 | IPR027417 | P-loop containing nucleoside triphosphate hydrolase |
| Pfam | PF00612 | 0.002 | 854 | 873 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 7.858 | 854 | 882 | IPR000048 | IQ motif, EF-hand binding site |
| SMART | SM00015 | 0.0092 | 875 | 897 | IPR000048 | IQ motif, EF-hand binding site |
| PROSITE profile | PS50096 | 9.304 | 876 | 900 | IPR000048 | IQ motif, EF-hand binding site |
| Pfam | PF00612 | 4.6E-4 | 878 | 897 | IPR000048 | IQ motif, EF-hand binding site |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0071275 | Biological Process | cellular response to aluminum ion | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0005515 | Molecular Function | protein binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1038 aa Download sequence Send to blast |
MADRGSFGFA PRLDIKQLLS EAQHRWLRPA EICEILRNHQ KFHIASEPPN RPPSGSLFLF 60 DRKVLRYFRK DGHNWRKKKD GKTVKEAHEK LKVGSIDVLH CYYAHGEDNE NFQRRCYWML 120 EQDLMHIVFV HYLEVKGNRM STGGTKENHS NSLSGTGSVN VDSTATRSSI LSPLCEDADS 180 GDSRQASSSL QPNPEPQTVV PQIIHHQNAI TMNSCNTASV LGNPDGWTSA PGIGLVSQVH 240 GNRVKESDSQ RSGDVPAWDA SFENSLARYQ NLPYNAPLTQ TQPLQNQVNW QIPVQESVPL 300 QKWPMDSHSG MTDTTDLALF GQRGHENFGT FSSLLGSQNQ QPSSFQAPFT NNEAAYIPKL 360 GPEDLIYEAS ANQTLPLRKA LLKKEDSLKK VDSFSRWVSK ELGEMEDLQM QSSSGGIAWT 420 SVECETAAAG SSLSPSLSED QRFTMIDFWP KWTQTDSEVE VMVIGTFLLS PQEVTSYSWS 480 CMFGEVEVPA EILVDGVLCC HAPPHEVGRV PFYITCSDRF SCSEVREFDF LPGSTRKLNA 540 TDIYGANTIE ASLHLRFDSL LALRSSVEEH HIFENVGEKR RKISKIMLLK DEKESLLPGT 600 VEKDLSALEA KERLIREEFE DKLYLWLIHK VTEEGKGPNI LDEDGQGVLH LAAALGYDWG 660 IKPILAAGVS INFRDANGWS ALHWAAFSGR EDTVAVLVSL GADAGALADP SPEHPLGKTA 720 ADLAYGNGHR GISGFLAESS LTSYLEKLTV DAKENSSADS SGAKAVLTVA ERTATPMSYG 780 DVPETLSMKD SLTAVLNATQ AADRLHQVFR MQSFQRKQLS ELGGDNEFDI SDELAVSFAA 840 AKTKKPGHSN GAVHAAAVQI QKKYRGWKKR KEFLLIRQRI VKIQAHVRGH QVRKQYRAII 900 WSVGLLEKII LRWRRKGSGL RGFKRDTITK PAEPVCPAPQ EDDYDFLKEG RKQTEERLQK 960 ALTRVKSMAQ YPEARAQYRR LLTVVEGFRE NEASSSSSAL KNNNSNTEEA ANYNEEDDLI 1020 DIDSLLDDDS FMSLAFE* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
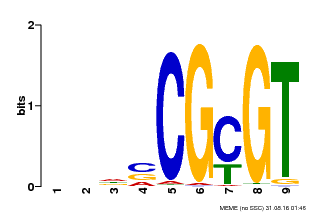
| UniProt | Transcription activator that binds to the DNA consensus sequence 5'-[ACG]CGCG[GTC]-3' (By similarity). Regulates transcriptional activity in response to calcium signals (Probable). Binds calmodulin in a calcium-dependent manner (By similarity). Involved in freezing tolerance in association with CAMTA1 and CAMTA3. Contributes together with CAMTA1 and CAMTA3 to the positive regulation of the cold-induced expression of DREB1A/CBF3, DREB1B/CBF1 and DREB1C/CBF2 (PubMed:23581962). Involved together with CAMTA3 and CAMTA4 in the positive regulation of a general stress response (PubMed:25039701). Involved in tolerance to aluminum. Binds to the promoter of ALMT1 transporter and contributes to the positive regulation of aluminum-induced expression of ALMT1 (PubMed:25627216). {ECO:0000250|UniProtKB:Q8GSA7, ECO:0000269|PubMed:23581962, ECO:0000269|PubMed:25039701, ECO:0000269|PubMed:25627216, ECO:0000305|PubMed:11925432}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00043 | PBM | Transfer from AT5G64220 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Csa11g101340.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By salt, wounding, abscisic acid, H(2)O(2) and salicylic acid (PubMed:12218065). Induced by aluminum (PubMed:25627216). {ECO:0000269|PubMed:12218065, ECO:0000269|PubMed:25627216}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK229403 | 0.0 | AK229403.1 Arabidopsis thaliana mRNA for Calmodulin-binding transcription activator 2, complete cds, clone: RAFL16-66-C08. | |||
| GenBank | BT010874 | 0.0 | BT010874.1 Arabidopsis thaliana At5g64220 gene, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010444352.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 2-like isoform X1 | ||||
| Refseq | XP_010444355.1 | 0.0 | PREDICTED: calmodulin-binding transcription activator 2-like isoform X3 | ||||
| Swissprot | Q6NPP4 | 0.0 | CMTA2_ARATH; Calmodulin-binding transcription activator 2 | ||||
| TrEMBL | D7MR88 | 0.0 | D7MR88_ARALL; Calmodulin-binding protein | ||||
| STRING | XP_010444355.1 | 0.0 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4082 | 24 | 52 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G64220.2 | 0.0 | Calmodulin-binding transcription activator protein with CG-1 and Ankyrin domains | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



