 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Csa11g102540.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Camelina
|
||||||||
| Family | AP2 | ||||||||
| Protein Properties | Length: 514aa MW: 57217.1 Da PI: 6.3936 | ||||||||
| Description | AP2 family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | AP2 | 33.1 | 1.4e-10 | 226 | 283 | 1 | 56 |
AP2 1 sgykGVrwdkkrgrWvAeIrd.pseng...krkrfslgkfgtaeeAakaaiaarkklege 56
s y+GV++++++gr++A+++d + rk + g ++ +++Aa+a++ a++k++g+
Csa11g102540.1 226 SIYRGVTRHRWTGRYEAHLWDnSCR-RegqARKGRQ-GGYDKEDKAARAYDLAALKYWGH 283
57*******************4444.2344336655.7799999*************997 PP
| |||||||
| 2 | AP2 | 49.1 | 1.4e-15 | 354 | 403 | 3 | 55 |
AP2 3 ykGVrwdkkrgrWvAeIrdpsengkrkrfslgkfgtaeeAakaaiaarkkleg 55
y+GV++++++grW A+I + +k +lg+f t+eeAa+a++ a+ k++g
Csa11g102540.1 354 YRGVTRHHQQGRWQARIGRVAG---NKDLYLGTFATEEEAAEAYDIAAIKFRG 403
9***************988532...5************************997 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF00847 | 1.7E-8 | 226 | 283 | IPR001471 | AP2/ERF domain |
| CDD | cd00018 | 5.24E-18 | 226 | 292 | No hit | No description |
| SuperFamily | SSF54171 | 3.86E-14 | 226 | 291 | IPR016177 | DNA-binding domain |
| SMART | SM00380 | 6.4E-20 | 227 | 296 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 3.5E-11 | 227 | 291 | IPR001471 | AP2/ERF domain |
| PROSITE profile | PS51032 | 16.95 | 227 | 290 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 7.6E-5 | 228 | 239 | IPR001471 | AP2/ERF domain |
| PRINTS | PR00367 | 7.6E-5 | 272 | 292 | IPR001471 | AP2/ERF domain |
| SuperFamily | SSF54171 | 1.05E-17 | 352 | 412 | IPR016177 | DNA-binding domain |
| CDD | cd00018 | 4.88E-11 | 352 | 411 | No hit | No description |
| PROSITE profile | PS51032 | 19.269 | 353 | 411 | IPR001471 | AP2/ERF domain |
| SMART | SM00380 | 3.3E-30 | 353 | 417 | IPR001471 | AP2/ERF domain |
| Gene3D | G3DSA:3.30.730.10 | 4.1E-18 | 353 | 411 | IPR001471 | AP2/ERF domain |
| Pfam | PF00847 | 6.9E-10 | 354 | 403 | IPR001471 | AP2/ERF domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0010492 | Biological Process | maintenance of shoot apical meristem identity | ||||
| GO:0060772 | Biological Process | leaf phyllotactic patterning | ||||
| GO:0060774 | Biological Process | auxin mediated signaling pathway involved in phyllotactic patterning | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 514 aa Download sequence Send to blast |
MAPPPMTNLL TFSLSPMEML KSPDQSHFPP PPSYDASSTP YLIDNFYVFK EEAAASSMAD 60 STTLSTFFDS QILSQTQIPK LEDFLGDSFV RYSDDQTETQ DDSSLIRLYD LRDHTVTGGV 120 TGFFSDHHHH HDFKTVNSSG SQILDDSASD IVATHLSGHA VKSSSTTAEL GFNGGASTGE 180 ALSLAVNNNT SDQPPQSNKK TTASTKKETS DDSKKKVVEP LEQRTSIYRG VTRHRWTGRY 240 EAHLWDNSCR REGQARKGRQ GGYDKEDKAA RAYDLAALKY WGHIATTNFP VASYSKEVEE 300 MNHMTKQEFX IATTNFPVAS YSKELEEMNH MTKQEFIASL RRKSSGFSRG ASMYRGVTRH 360 HQQGRWQARI GRVAGNKDLY LGTFATEEEA AEAYDIAAIK FRGINAVTNF EMNRYDVEAV 420 MKCSFPVGGA AKRHKVAVES PPSSDHSLQQ QDLLPSSSSS DVNPNSIPCG IPFEPSLLYH 480 HQNFFQHYPL VSDSTVQVPM NPAEFFLWPN QSY* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Probably acts as a transcriptional activator. Binds to the GCC-box pathogenesis-related promoter element. May be involved in the regulation of gene expression by stress factors and by components of stress signal transduction pathways (By similarity). {ECO:0000250}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00589 | DAP | Transfer from AT5G65510 | Download |
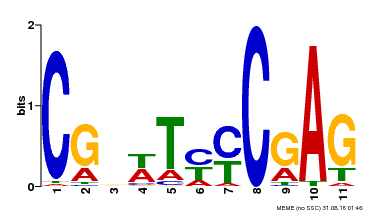
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Csa11g102540.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AY560887 | 0.0 | AY560887.1 Arabidopsis thaliana putative AP2/EREBP transcription factor (At5g65510) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_019088688.1 | 0.0 | PREDICTED: AP2-like ethylene-responsive transcription factor AIL7 | ||||
| Swissprot | Q6J9N8 | 0.0 | AIL7_ARATH; AP2-like ethylene-responsive transcription factor AIL7 | ||||
| TrEMBL | R0EWM6 | 0.0 | R0EWM6_9BRAS; Uncharacterized protein | ||||
| STRING | XP_010444513.1 | 0.0 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3333 | 27 | 65 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G65510.1 | 0.0 | AINTEGUMENTA-like 7 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



