 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Csa14g026180.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Camelina
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 1051aa MW: 115918 Da PI: 8.3146 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 129.9 | 9.4e-41 | 138 | 215 | 1 | 78 |
--SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 1 lCqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
+Cqv++C++dls+ak+yhrrhkvCevhska+++lv +++qrfCqqCsrfh lsefDe+krsCrrrLa+hn+rrrk+++
Csa14g026180.1 138 MCQVDNCTEDLSHAKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHLLSEFDEGKRSCRRRLAGHNRRRRKTTQ 215
6**************************************************************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 6.4E-34 | 133 | 200 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.077 | 136 | 213 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 3.66E-37 | 137 | 214 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 9.3E-29 | 139 | 212 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51296 | 9.638 | 628 | 689 | IPR017941 | Rieske [2Fe-2S] iron-sulphur domain |
| Gene3D | G3DSA:1.25.40.20 | 2.7E-7 | 802 | 947 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 4.56E-10 | 802 | 945 | No hit | No description |
| SuperFamily | SSF48403 | 3.55E-8 | 806 | 948 | IPR020683 | Ankyrin repeat-containing domain |
| PROSITE profile | PS50297 | 8.8 | 886 | 958 | IPR020683 | Ankyrin repeat-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0055114 | Biological Process | oxidation-reduction process | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0005886 | Cellular Component | plasma membrane | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0016491 | Molecular Function | oxidoreductase activity | ||||
| GO:0051537 | Molecular Function | 2 iron, 2 sulfur cluster binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 1051 aa Download sequence Send to blast |
MDEVGAQVAT PMFIHPSLSP MGRKRDLYYP MSSRLVPSQQ QQPQRRDEWN SKMWDWDSRR 60 FEAKPVDAQT LSLGNVTTQQ FDLTSRNKSG GGEERGLDLN LGSGLTALEE TTTTTTAQNV 120 RPSKKVRSGS PGGGNYPMCQ VDNCTEDLSH AKDYHRRHKV CEVHSKATKA LVGKQMQRFC 180 QQCSRFHLLS EFDEGKRSCR RRLAGHNRRR RKTTQPEEIA SGVVVPGNRD NNTSNANMDL 240 MALLTALACA QGKNDVKPVS SPAVPDREQL LQILNKINAL PLPMDLVSKL NNIGSLARKN 300 LDQPMANPTN DMNGASPSTM DLLAVLSSTL GSSSPDALAI LSQGRFGNKD SDKTKLSSYD 360 HGVTTNVEKR TFGFSSGGGE RSSSSNQSPS QDSDSRAQDT RSSLSLQLFT SSPEDESRPT 420 VASSRKYYSS ASSNPVEDRS PSSSPVMQEL FPLQTSPETM RSNNHNNSSP SRTGCLPLEL 480 FGASNRGTAN PNFKGFGQQS GYASSGSDYS PPSLNSDAQD RTGKIVFKLL DKDPSQLPGT 540 LRSEIYNWLS NIPSEMESYI RPGCVVLSVY IAMSPAAWEQ LERNLLQRLG VLLQNPQSDF 600 WRNVRFIVNT GRQLASHKNG RVRCSKSWRT WNSPELLSVS PVAVVAGAET SLVVRGRSLT 660 NDGISIRCTH MGSYMSMEVT GAACRQAVCD ELNVNSFKVQ NAQPGFLGRC FIEVENGFRG 720 DSFPLIIANA SICKELNRLE EEFHPKSQDT SEEQAQSTDS RPTSREEVLC FLNELGWLFQ 780 KNQTSELREQ SDFSLARFKF LLVCSVERDY CALIRTLLDM LVERNLVNDE LNTEASDMLA 840 EIQLLNRAVK RKSTKMVELL IHYTVKSSAL DSSNNFVFLP NITGPGGITP LHLAACTSGS 900 DDMVDLLTND PQEIGLSSWN SLRDATGQTP YSYAAIRNNH SYNSLVARKL ADKRNKQVSL 960 NIENEIVDQK GLSKRLSSEM NKSSCASCAT VALKYQRRVS GSHRLFPTPI IHSMLAVATV 1020 CVCVCVFMHA FPIVRQGSHF SWGGLDYGSI * |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1ul4_A | 4e-32 | 129 | 212 | 1 | 84 | squamosa promoter binding protein-like 4 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3' (By similarity). May play a role in plant development. {ECO:0000250, ECO:0000269|PubMed:15703061}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00155 | DAP | Transfer from AT1G20980 | Download |
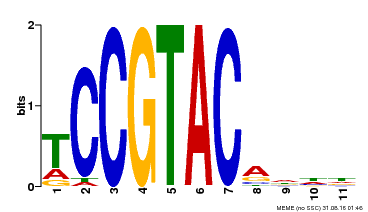
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Csa14g026180.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK226907 | 0.0 | AK226907.1 Arabidopsis thaliana mRNA for squamosa promoter binding protein-like 14, clone: RAFL09-16-H09. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010459792.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 14 | ||||
| Swissprot | Q8RY95 | 0.0 | SPL14_ARATH; Squamosa promoter-binding-like protein 14 | ||||
| TrEMBL | R0GUT0 | 0.0 | R0GUT0_9BRAS; Uncharacterized protein | ||||
| STRING | XP_010459792.1 | 0.0 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3999 | 25 | 54 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G20980.1 | 0.0 | squamosa promoter binding protein-like 14 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



