 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Csa16g005930.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Camelina
|
||||||||
| Family | SBP | ||||||||
| Protein Properties | Length: 905aa MW: 101114 Da PI: 6.3469 | ||||||||
| Description | SBP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | SBP | 132.1 | 1.9e-41 | 117 | 193 | 2 | 78 |
-SSTT-----TT--HHHHHTT--HHHHT-S-EEETTEEEEE-TTTSSEEETTT--SS--S-STTTT-------S--- CS
SBP 2 CqvegCeadlseakeyhrrhkvCevhskapvvlvsgleqrfCqqCsrfhelsefDeekrsCrrrLakhnerrrkkqa 78
Cqve+C adl++ak+yhrrhkvCe+hska+++lv ++qrfCqqCsrfh l+efDe+krsCrrrLa+hn+rrrk+++
Csa16g005930.1 117 CQVENCGADLTKAKDYHRRHKVCEMHSKATSALVGSIMQRFCQQCSRFHVLEEFDEGKRSCRRRLAGHNKRRRKANP 193
**************************************************************************875 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.1100.10 | 1.2E-32 | 113 | 178 | IPR004333 | Transcription factor, SBP-box |
| PROSITE profile | PS51141 | 32.077 | 114 | 191 | IPR004333 | Transcription factor, SBP-box |
| SuperFamily | SSF103612 | 1.57E-38 | 116 | 196 | IPR004333 | Transcription factor, SBP-box |
| Pfam | PF03110 | 6.3E-30 | 117 | 190 | IPR004333 | Transcription factor, SBP-box |
| Gene3D | G3DSA:1.25.40.20 | 2.7E-6 | 694 | 797 | IPR020683 | Ankyrin repeat-containing domain |
| SuperFamily | SSF48403 | 2.18E-7 | 695 | 796 | IPR020683 | Ankyrin repeat-containing domain |
| CDD | cd00204 | 8.07E-8 | 698 | 796 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 905 aa Download sequence Send to blast |
MEARRTQGGE EEGHSLYGFS GKTSVAEWDL NDWKWDGHLF VARQLNHGSS NTSSACSDEA 60 TVVDIMDNMN KRRAVTVIPM DDDDAPKLTL NLGGCGSGNF AKKTKLGGAL QTRAISCQVE 120 NCGADLTKAK DYHRRHKVCE MHSKATSALV GSIMQRFCQQ CSRFHVLEEF DEGKRSCRRR 180 LAGHNKRRRK ANPDTLGDGT PTSDGQTSNY LLITLLKILS NIHLNQSDQT GDQLLSHLLK 240 SLVSQPGEHV GKDLVGLMQG GGGLEASQAL LSLEQAHRED IKHHSVNVLE TPLQEVYVNG 300 SQERAVLDKS QKQVKMNAFD LNDVYIDSDD TTDIERSSPP PTNPATSSLD YHQDSRQSSP 360 PQTSRNSDSA SDQSPSSSSR DAPSRTDRIV FKLFGKVPND FPVALRGQIL DWLAHTPTDM 420 ESYIRPGCIV LTVYLRQDEA SWEELCCDLS FSLRRLLDLS HDPLWTDGWI YLRVQNQFAF 480 AFNGQVVLDT SLPLRSHDNS QIITVRPLAV TRKAQFTVKG INLHRPGTRV LCAVEGTYLV 540 QEETQGVTEE SDDLKEQNKP DSVKFSCEIP IGSGRGFMEI EDQDGLSSSF FPFIVSEDED 600 VCSEIQRLES TLEFTGTDSP MQAMYFIHEI GWLLHKSELS SRFAESDHLF PLIRFKFLIE 660 FSMDREWCAV IKKLLNILFE EGTVDLSPDA ALSELCLLHR AVRKNSKLMV EMLLRFIPKK 720 TNLALSGLFR PDAAGPSGLT PLHIAAGKDG SEDVLDALTD DPGMIGIQAW KTSRDNTGFT 780 PEDYARLRGH FSYIHLVQRK LNRKPTAEEH VVVNIPESFN IDHKQEKRSL KDSSSLKINQ 840 CKLCDHKRVF VTRQHSSVAY RPAMLSMVAI AAVCVCVALL FKSCPEVLYV FQPFRWELLA 900 YGTS* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1wj0_A | 2e-31 | 114 | 171 | 3 | 60 | squamosa promoter-binding protein-like 12 |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Trans-acting factor that binds specifically to the consensus nucleotide sequence 5'-TNCGTACAA-3'. {ECO:0000269|PubMed:16554053}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00060 | PBM | Transfer from AT3G60030 | Download |
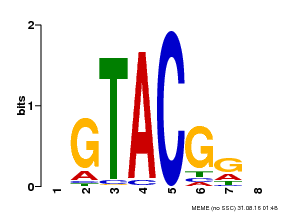
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Csa16g005930.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AJ132096 | 0.0 | AJ132096.1 Arabidopsis thaliana mRNA for squamosa promoter binding protein-like 12. | |||
| GenBank | BT003817 | 0.0 | BT003817.1 Arabidopsis thaliana clone C105276 putative squamosa promoter binding protein 12 (At3g60030) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010469241.1 | 0.0 | PREDICTED: squamosa promoter-binding-like protein 12 | ||||
| Swissprot | Q9S7P5 | 0.0 | SPL12_ARATH; Squamosa promoter-binding-like protein 12 | ||||
| TrEMBL | D7LWK1 | 0.0 | D7LWK1_ARALL; Uncharacterized protein | ||||
| STRING | XP_010469241.1 | 0.0 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM3030 | 27 | 64 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G60030.1 | 0.0 | squamosa promoter-binding protein-like 12 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



