 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Csa16g048760.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Camelina
|
||||||||
| Family | MYB | ||||||||
| Protein Properties | Length: 299aa MW: 34157.6 Da PI: 4.9988 | ||||||||
| Description | MYB family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 51.9 | 1.7e-16 | 16 | 63 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg+W++eEd++l+ ++++G ++W++ ++ g+ R++k+c++rw +yl
Csa16g048760.1 16 RGPWSPEEDLKLISFIQKFGHENWRSLPKQSGLLRCGKSCRLRWINYL 63
89******************************99************97 PP
| |||||||
| 2 | Myb_DNA-binding | 55 | 1.8e-17 | 69 | 114 | 1 | 48 |
TSSS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHHT CS
Myb_DNA-binding 1 rgrWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqkyl 48
rg++T+eE+e ++++++ +G++ W++Ia+ ++ gRt++++k+ w+++l
Csa16g048760.1 69 RGNFTAEEEETIIKLHQNYGNK-WSKIASQLP-GRTDNEIKNVWHTHL 114
89********************.*********.************986 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 1.5E-24 | 7 | 66 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 15.411 | 11 | 63 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 3.67E-30 | 14 | 110 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 2.4E-13 | 15 | 65 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 5.1E-15 | 16 | 63 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 2.36E-9 | 18 | 63 | No hit | No description |
| PROSITE profile | PS51294 | 26.4 | 64 | 118 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 3.0E-28 | 67 | 119 | IPR009057 | Homeodomain-like |
| SMART | SM00717 | 3.2E-15 | 68 | 116 | IPR001005 | SANT/Myb domain |
| Pfam | PF00249 | 6.9E-16 | 69 | 114 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 4.50E-10 | 71 | 114 | No hit | No description |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0009751 | Biological Process | response to salicylic acid | ||||
| GO:0009753 | Biological Process | response to jasmonic acid | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:2000652 | Biological Process | regulation of secondary cell wall biogenesis | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 299 aa Download sequence Send to blast |
MGKGRAPCCD KTKVKRGPWS PEEDLKLISF IQKFGHENWR SLPKQSGLLR CGKSCRLRWI 60 NYLRPDLKRG NFTAEEEETI IKLHQNYGNK WSKIASQLPG RTDNEIKNVW HTHLKKRLVQ 120 SDLTADEPAS SPCSSDYVSR GNEKKSRAED SFNRTTNHES SSRTSAVSSG GSINHSNQED 180 DPKIGLTFEY IEEAYSEFKD IIQEVDKSDM LEIPFDTDTD IWSYLDASNS FQQCAANEFS 240 SASRAEEESD EDEVKKWLKH LESELGLEED DSQQQRTQDN KEESSSLLKT SCYELMIH* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1gv2_A | 4e-25 | 16 | 118 | 4 | 105 | MYB PROTO-ONCOGENE PROTEIN |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
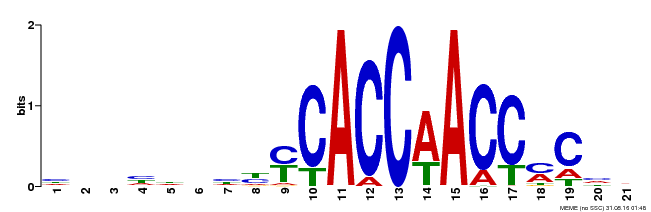
| UniProt | Transcriptional activator that binds DNA to the AC cis-elements 5'-ACCTACC-3', 5'-ACCAACC-3' and 5'-ACCTAAC-3' of promoters and specifically activates lignin biosynthetic genes during secondary wall formation mediated by SND1. {ECO:0000269|PubMed:19122102}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00249 | DAP | Transfer from AT1G79180 | Download |
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Csa16g048760.1 |
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Slightly induced by UV light (PubMed:9839469). Triggered by salicylic acid and jasmonic acid (PubMed:16463103). Regulated by the SND1 close homologs NST1, NST2, VND6, and VND7 and their downstream targets MYB46 and MYB83 (PubMed:19122102, PubMed:22197883). {ECO:0000269|PubMed:16463103, ECO:0000269|PubMed:19122102, ECO:0000269|PubMed:22197883, ECO:0000269|PubMed:9839469}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK175344 | 1e-161 | AK175344.1 Arabidopsis thaliana mRNA for putative transcription factor (MYB63), complete cds, clone: RAFL21-78-M22. | |||
| GenBank | AY519572 | 1e-161 | AY519572.1 Arabidopsis thaliana MYB transcription factor (At1g79180) mRNA, complete cds. | |||
| GenBank | BT033125 | 1e-161 | BT033125.1 Arabidopsis thaliana unknown protein (At1g79180) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010472758.1 | 0.0 | PREDICTED: myb-related protein Myb4-like isoform X2 | ||||
| Swissprot | Q6R0A6 | 1e-157 | MYB63_ARATH; Transcription factor MYB63 | ||||
| TrEMBL | R0GID1 | 1e-170 | R0GID1_9BRAS; Uncharacterized protein | ||||
| STRING | XP_010472758.1 | 0.0 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM4 | 28 | 2646 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G79180.1 | 1e-138 | myb domain protein 63 | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



