 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Csa18g032080.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; malvids; Brassicales; Brassicaceae; Camelineae; Camelina
|
||||||||
| Family | MYB_related | ||||||||
| Protein Properties | Length: 292aa MW: 31713.3 Da PI: 6.6283 | ||||||||
| Description | MYB_related family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | Myb_DNA-binding | 45.9 | 1.3e-14 | 145 | 189 | 3 | 47 |
SS-HHHHHHHHHHHHHTTTT-HHHHHHHHTTTS-HHHHHHHHHHH CS
Myb_DNA-binding 3 rWTteEdellvdavkqlGggtWktIartmgkgRtlkqcksrwqky 47
+WT+eE++l++ + k++G+g+W+ I+r + +Rt+ q+ s+ qky
Csa18g032080.1 145 PWTEEEHKLFLMGLKKYGKGDWRNISRNFVITRTPTQVASHAQKY 189
8*****************************99************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51293 | 7.409 | 31 | 86 | IPR017884 | SANT domain |
| SMART | SM00717 | 1.4E-7 | 32 | 84 | IPR001005 | SANT/Myb domain |
| CDD | cd00167 | 5.54E-7 | 35 | 81 | No hit | No description |
| SuperFamily | SSF46689 | 8.25E-10 | 35 | 88 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 20.772 | 138 | 194 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 7.25E-17 | 140 | 193 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.8E-17 | 141 | 193 | IPR006447 | Myb domain, plants |
| SMART | SM00717 | 3.2E-13 | 142 | 192 | IPR001005 | SANT/Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 1.2E-12 | 142 | 189 | IPR009057 | Homeodomain-like |
| CDD | cd00167 | 2.11E-11 | 145 | 190 | No hit | No description |
| Pfam | PF00249 | 5.0E-13 | 145 | 189 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 292 aa Download sequence Send to blast |
MEVLRPTTSH VSGGNWLMED TKSNAGVAAS GEGCTWTAAE NKAFENALAV YDDNTPDRWQ 60 KVAAVIPGKT VSDVIRQYND LEADVSSIEA GLIPVPGYVT SPFTLDWAGG GGSGGCNGFK 120 PGHSVCYKRS SAGRSPELER KKGVPWTEEE HKLFLMGLKK YGKGDWRNIS RNFVITRTPT 180 QVASHAQKYF IRQLSGGKDK RRASIHDITT VNLEDEASLE SNKSSIVVGE QRSRLTLFPW 240 NETDNNGTHA DAFNITIGNA ISGVHSYGQV MLGGFNNADS CYDAQNTMFQ L* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 2cjj_A | 4e-18 | 28 | 101 | 3 | 76 | RADIALIS |
| Search in ModeBase | ||||||
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in the dorsovental asymmetry of flowers. Promotes ventral identity. {ECO:0000269|PubMed:11937495, ECO:0000269|PubMed:9118809}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00568 | DAP | Transfer from AT5G58900 | Download |
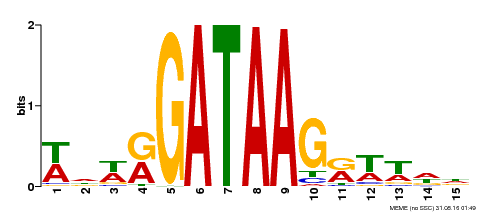
| |||
| Cis-element ? help Back to Top | |
|---|---|
| Source | Link |
| PlantRegMap | Csa18g032080.1 |
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | AK118891 | 0.0 | AK118891.1 Arabidopsis thaliana At5g58900 mRNA for putative I-box binding factor, complete cds, clone: RAFL21-22-M07. | |||
| GenBank | AY519533 | 0.0 | AY519533.1 Arabidopsis thaliana MYB transcription factor (At5g58900) mRNA, complete cds. | |||
| GenBank | BT005473 | 0.0 | BT005473.1 Arabidopsis thaliana clone U51211 putative I-box binding factor (At5g58900) mRNA, complete cds. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_010483499.1 | 0.0 | PREDICTED: transcription factor DIVARICATA-like isoform X1 | ||||
| Swissprot | Q8S9H7 | 1e-104 | DIV_ANTMA; Transcription factor DIVARICATA | ||||
| TrEMBL | R0GRH6 | 0.0 | R0GRH6_9BRAS; Uncharacterized protein | ||||
| STRING | XP_010483499.1 | 0.0 | (Camelina sativa) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Malvids | OGEM1228 | 27 | 100 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G58900.1 | 0.0 | Homeodomain-like transcriptional regulator | ||||
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



