 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.043300.2 | ||||||||
| Common Name | Csa_6G504650, LOC101207689 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | SRS | ||||||||
| Protein Properties | Length: 326aa MW: 35200.4 Da PI: 7.1334 | ||||||||
| Description | SRS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF702 | 229.5 | 6.1e-71 | 105 | 261 | 3 | 154 |
DUF702 3 sgtasCqdCGnqakkdCaheRCRtCCksrgfdCathvkstWvpaakrrerqqqlaaasskaaasa........aeaaskrkrelkskkqsalsst 89
g++sCqdCGnqakkdC+h+RCRtCCksrgf+C+thvkstWvpaakrrerq++laa+++++ +++ ++++kr+r ++s+l++t
Cucsa.043300.2 105 GGGISCQDCGNQAKKDCSHMRCRTCCKSRGFHCETHVKSTWVPAAKRRERQDKLAALQTHHHHHHqqqlqlhgGDNNPKRHR---DYNSSSLACT 196
5789****************************************************99988877767777544444444444...569999**** PP
DUF702 90 klssaeskkeletsslPeevsseavfrcvrvssvddgeeelaYqtavsigGhvfkGiLydqGlee 154
++++++++ le+ ++P+e++s+avfrcvrvss dd ++++aYqtav+igGhvfkGiLydqG+e+
Cucsa.043300.2 197 LIPTNNNTSGLEIGNFPAELNSPAVFRCVRVSSADDTDDQYAYQTAVNIGGHVFKGILYDQGPEN 261
***************************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05142 | 1.3E-66 | 107 | 260 | IPR007818 | Protein of unknown function DUF702 |
| TIGRFAMs | TIGR01623 | 1.1E-27 | 109 | 151 | IPR006510 | Zinc finger, lateral root primordium type 1 |
| TIGRFAMs | TIGR01624 | 2.3E-27 | 212 | 259 | IPR006511 | Lateral Root Primordium type 1, C-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010051 | Biological Process | xylem and phloem pattern formation | ||||
| GO:0010252 | Biological Process | auxin homeostasis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048479 | Biological Process | style development | ||||
| GO:0048480 | Biological Process | stigma development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 326 aa Download sequence Send to blast |
MSGFFTLGGG GGGRDEEQNN NRPNQFIHPP PDSSLFWYKA GNVELWQQQQ QQQHHHHHQE 60 QPLFQTQSRT TTTTTPYSAP LAVRNSDESS SRSGNFMMIS SGSGGGGISC QDCGNQAKKD 120 CSHMRCRTCC KSRGFHCETH VKSTWVPAAK RRERQDKLAA LQTHHHHHHQ QQLQLHGGDN 180 NPKRHRDYNS SSLACTLIPT NNNTSGLEIG NFPAELNSPA VFRCVRVSSA DDTDDQYAYQ 240 TAVNIGGHVF KGILYDQGPE NNYIPPGETS SGGGGSSSGV QPLNFIAGAA DATTGSGGST 300 AALPLLDPSS LYSTPLNSFM AGDYV* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds DNA on 5'-ACTCTAC-3' and promotes auxin homeostasis-regulating gene expression (e.g. YUC genes), as well as genes affecting stamen development, cell expansion and timing of flowering. Synergistically with other SHI-related proteins, regulates gynoecium, stamen and leaf development in a dose-dependent manner, controlling apical-basal patterning. Promotes style and stigma formation, and influences vascular development during gynoecium development. May also have a role in the formation and/or maintenance of the shoot apical meristem (SAM). Suppressor of the gibberellin (GA) signal transduction. {ECO:0000269|PubMed:10368174, ECO:0000269|PubMed:11706176, ECO:0000269|PubMed:16740146}. | |||||
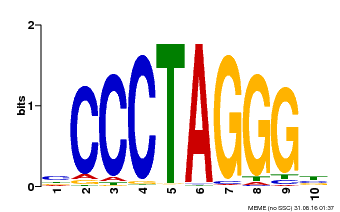
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00613 | PBM | Transfer from AT3G51060 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681875 | 0.0 | LN681875.1 Cucumis melo genomic scaffold, anchoredscaffold00007. | |||
| GenBank | LN713262 | 0.0 | LN713262.1 Cucumis melo genomic chromosome, chr_8. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004149998.1 | 0.0 | PREDICTED: protein SHI RELATED SEQUENCE 1 | ||||
| Swissprot | Q9XGX0 | 2e-78 | SHI_ARATH; Protein SHORT INTERNODES | ||||
| TrEMBL | A0A0A0KGL4 | 0.0 | A0A0A0KGL4_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004172666.1 | 0.0 | (Cucumis sativus) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G66350.1 | 1e-67 | Lateral root primordium (LRP) protein-related | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.043300.2 |
| Entrez Gene | 101207689 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



