 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.043300.3 | ||||||||
| Common Name | Csa_6G504650, LOC101207689 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | SRS | ||||||||
| Protein Properties | Length: 236aa MW: 25033.7 Da PI: 7.7784 | ||||||||
| Description | SRS family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | DUF702 | 230.7 | 2.5e-71 | 9 | 165 | 3 | 154 |
DUF702 3 sgtasCqdCGnqakkdCaheRCRtCCksrgfdCathvkstWvpaakrrerqqqlaaasskaaasa........aeaaskrkrelkskkqsalsst 89
g++sCqdCGnqakkdC+h+RCRtCCksrgf+C+thvkstWvpaakrrerq++laa+++++ +++ ++++kr+r ++s+l++t
Cucsa.043300.3 9 GGGISCQDCGNQAKKDCSHMRCRTCCKSRGFHCETHVKSTWVPAAKRRERQDKLAALQTHHHHHHqqqlqlhgGDNNPKRHR---DYNSSSLACT 100
5789****************************************************99988877767777544444444444...569999**** PP
DUF702 90 klssaeskkeletsslPeevsseavfrcvrvssvddgeeelaYqtavsigGhvfkGiLydqGlee 154
++++++++ le+ ++P+e++s+avfrcvrvss dd ++++aYqtav+igGhvfkGiLydqG+e+
Cucsa.043300.3 101 LIPTNNNTSGLEIGNFPAELNSPAVFRCVRVSSADDTDDQYAYQTAVNIGGHVFKGILYDQGPEN 165
***************************************************************97 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF05142 | 5.2E-67 | 11 | 164 | IPR007818 | Protein of unknown function DUF702 |
| TIGRFAMs | TIGR01623 | 6.8E-28 | 13 | 55 | IPR006510 | Zinc finger, lateral root primordium type 1 |
| TIGRFAMs | TIGR01624 | 1.4E-27 | 116 | 163 | IPR006511 | Lateral Root Primordium type 1, C-terminal |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0010051 | Biological Process | xylem and phloem pattern formation | ||||
| GO:0010252 | Biological Process | auxin homeostasis | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:0048479 | Biological Process | style development | ||||
| GO:0048480 | Biological Process | stigma development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 236 aa Download sequence Send to blast |
MMISSGSGGG GISCQDCGNQ AKKDCSHMRC RTCCKSRGFH CETHVKSTWV PAAKRRERQD 60 KLAALQTHHH HHHQQQLQLH GGDNNPKRHR DYNSSSLACT LIPTNNNTSG LEIGNFPAEL 120 NSPAVFRCVR VSSADDTDDQ YAYQTAVNIG GHVFKGILYD QGPENNYIPP GETSSGGGGS 180 SSGVQPLNFI AGAADATTGS GGSTAALPLL DPSSLYSTPL NSFMAGTQFF LPPRS* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription activator that binds DNA on 5'-ACTCTAC-3' and promotes auxin homeostasis-regulating gene expression (e.g. YUC genes), as well as genes affecting stamen development, cell expansion and timing of flowering. Synergistically with other SHI-related proteins, regulates gynoecium, stamen and leaf development in a dose-dependent manner, controlling apical-basal patterning. Promotes style and stigma formation, and influences vascular development during gynoecium development. May also have a role in the formation and/or maintenance of the shoot apical meristem (SAM). {ECO:0000269|PubMed:12361963, ECO:0000269|PubMed:16740145, ECO:0000269|PubMed:16740146, ECO:0000269|PubMed:18811619, ECO:0000269|PubMed:20154152, ECO:0000269|PubMed:22318676}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00613 | PBM | Transfer from AT3G51060 | Download |
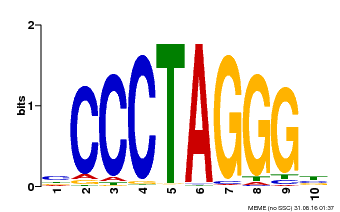
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Regulated by ESR1 and ESR2. {ECO:0000269|PubMed:21976484}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681875 | 0.0 | LN681875.1 Cucumis melo genomic scaffold, anchoredscaffold00007. | |||
| GenBank | LN713262 | 0.0 | LN713262.1 Cucumis melo genomic chromosome, chr_8. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004149998.1 | 1e-175 | PREDICTED: protein SHI RELATED SEQUENCE 1 | ||||
| Swissprot | Q9SD40 | 2e-79 | SRS1_ARATH; Protein SHI RELATED SEQUENCE 1 | ||||
| TrEMBL | A0A0A0KGL4 | 1e-174 | A0A0A0KGL4_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004172666.1 | 1e-175 | (Cucumis sativus) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT3G51060.1 | 1e-72 | Lateral root primordium (LRP) protein-related | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.043300.3 |
| Entrez Gene | 101207689 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



