 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.053540.2 | ||||||||
| Common Name | Csa_3G391380, LOC101219851 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 386aa MW: 42717 Da PI: 7.2382 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 40.2 | 6.1e-13 | 241 | 287 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCC.C...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPk.askapskKlsKaeiLekAveYIksLq 55
+h e+Er+RR+++N++f Lr ++P+ + K++Ka+ L A+ YI +Lq
Cucsa.053540.2 241 NHVEAERQRREKLNQRFYALRAVVPNiS------KMDKASLLGDAITYITDLQ 287
799***********************66......*****************99 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Pfam | PF14215 | 2.3E-20 | 16 | 113 | IPR025610 | Transcription factor MYC/MYB N-terminal |
| PROSITE profile | PS50888 | 16.831 | 237 | 286 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 6.02E-18 | 240 | 300 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 9.69E-14 | 240 | 291 | No hit | No description |
| Gene3D | G3DSA:4.10.280.10 | 4.5E-18 | 241 | 298 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Pfam | PF00010 | 2.0E-10 | 241 | 287 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 6.2E-14 | 243 | 292 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS51671 | 8.535 | 319 | 385 | IPR002912 | ACT domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0008152 | Biological Process | metabolic process | ||||
| GO:0016597 | Molecular Function | amino acid binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 386 aa Download sequence Send to blast |
MLQSSFGRSD EDGYGARRDE ASDIEMLYLT SKYYKFMCDS GSSLGESYKS GKSIWASDVT 60 SCLRNYQSRG FLAKVAGLQT LVFVPVKLGV VELGSTKSIP EDQGVLELVR ASFGGSITAQ 120 LKAFPRIFGH ELSLGGTKPR SLSINFSPKL EDDTNFSSEG YELQGLGGNH IFGNSSNGCR 180 GDDNDAKMFP HGNQEVVGGF NAQTRLSTME FPRDESSPQG DDRKPRKRGR KPANGREEPL 240 NHVEAERQRR EKLNQRFYAL RAVVPNISKM DKASLLGDAI TYITDLQMKI KVMETEKQIA 300 SGREKNTEID FHAREEDAVV RVSCPLDLHP VSKVIKTFRE HQIEAQESNV TTSTDNDKVI 360 HSFFIRTEGG AAEQLKEKLV AALSK* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5gnj_A | 5e-29 | 235 | 297 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_B | 5e-29 | 235 | 297 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_E | 5e-29 | 235 | 297 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_F | 5e-29 | 235 | 297 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_G | 5e-29 | 235 | 297 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_I | 5e-29 | 235 | 297 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_M | 5e-29 | 235 | 297 | 2 | 64 | Transcription factor MYC2 |
| 5gnj_N | 5e-29 | 235 | 297 | 2 | 64 | Transcription factor MYC2 |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 222 | 230 | RKPRKRGRK |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that negatively regulates jasmonate (JA) signaling (PubMed:30610166). Negatively regulates JA-dependent response to wounding, JA-induced expression of defense genes, JA-dependent responses against herbivorous insects, and JA-dependent resistance against Botrytis cinerea infection (PubMed:30610166). Plays a positive role in resistance against the bacterial pathogen Pseudomonas syringae pv tomato DC3000 (PubMed:30610166). {ECO:0000269|PubMed:30610166}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00099 | PBM | Transfer from AT4G16430 | Download |
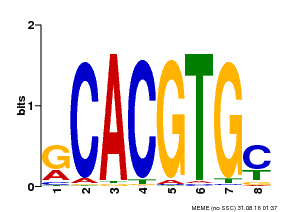
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by wounding, feeding with herbivorous insects, infection with the fungal pathogen Botrytis cinerea and infection with the bacterial pathogen Pseudomonas syringae pv tomato DC3000. {ECO:0000269|PubMed:30610166}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681835 | 0.0 | LN681835.1 Cucumis melo genomic scaffold, anchoredscaffold00064. | |||
| GenBank | LN713258 | 0.0 | LN713258.1 Cucumis melo genomic chromosome, chr_4. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011651443.1 | 0.0 | PREDICTED: transcription factor bHLH3 | ||||
| Swissprot | A0A3Q7H216 | 1e-143 | MTB3_SOLLC; Transcription factor MTB3 | ||||
| TrEMBL | A0A0A0L7H2 | 0.0 | A0A0A0L7H2_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004151980.1 | 0.0 | (Cucumis sativus) | ||||
| STRING | XP_004162392.1 | 0.0 | (Cucumis sativus) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G16430.1 | 1e-128 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.053540.2 |
| Entrez Gene | 101219851 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



