 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.087530.1 | ||||||||
| Common Name | Csa_2G408430, LOC101212135 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 454aa MW: 49597.6 Da PI: 6.1426 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 89.2 | 3.7e-28 | 169 | 223 | 1 | 56 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRla 56
k +++WtpeLH+rFv+aveqL G +kA+P++ilelm++++Lt+++v+SHLQkYR++
Cucsa.087530.1 169 KVKVDWTPELHRRFVQAVEQL-GVDKAVPSRILELMGIECLTRHNVASHLQKYRSH 223
5689*****************.********************************86 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| PROSITE profile | PS51294 | 17.242 | 166 | 225 | IPR017930 | Myb domain |
| Gene3D | G3DSA:1.10.10.60 | 6.0E-27 | 167 | 226 | IPR009057 | Homeodomain-like |
| SuperFamily | SSF46689 | 3.4E-18 | 167 | 226 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 1.9E-26 | 169 | 223 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 5.0E-8 | 172 | 221 | IPR001005 | SANT/Myb domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007165 | Biological Process | signal transduction | ||||
| GO:0009658 | Biological Process | chloroplast organization | ||||
| GO:0009910 | Biological Process | negative regulation of flower development | ||||
| GO:0010380 | Biological Process | regulation of chlorophyll biosynthetic process | ||||
| GO:0010638 | Biological Process | positive regulation of organelle organization | ||||
| GO:0045893 | Biological Process | positive regulation of transcription, DNA-templated | ||||
| GO:1900056 | Biological Process | negative regulation of leaf senescence | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 454 aa Download sequence Send to blast |
MLALSPIRSG NKDEKQGEME RFSIGGDDFP DFDDDTNLLD SINFDDLFVG INDGDVLPDL 60 EMDPELLAEF SVSGGEESEV NASVSLEKFD DNTLIGNKDN DDDEDQKDFD FRSSSQVVDQ 120 EILSKREDEL ATPTNVIEVS PLVKDGGDKS IKPLKASSSQ SKNSQGKRKV KVDWTPELHR 180 RFVQAVEQLG VDKAVPSRIL ELMGIECLTR HNVASHLQKY RSHRKHLLAR EAEAASWSQR 240 RQMYGGGGGG GGGGKREVSP WGAPPTMGFP PMTPMHPHFR PLHVWGHPPA MDQSLLHVWP 300 KHLPHSPSPP PPPPTPPPSS WPHTAAPPPP PDSSYWHHHH HHQRVPNGLT SGTPCFPQPI 360 PTTRFGGASF SVIPPPHPMY KAAEPTTSIG RSPTHPPLDS YPSKESIDSA IGDVLAKPWL 420 PLPLGLKPPS LDSVKVELQR QGVPKIPPST CAA* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1irz_A | 1e-15 | 166 | 221 | 2 | 57 | ARR10-B |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00022 | PBM | Transfer from AT2G20570 | Download |
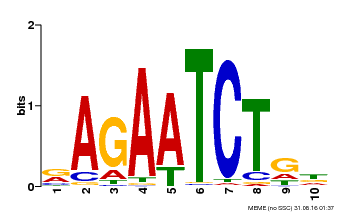
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681916 | 0.0 | LN681916.1 Cucumis melo genomic scaffold, anchoredscaffold00037. | |||
| GenBank | LN713265 | 0.0 | LN713265.1 Cucumis melo genomic chromosome, chr_11. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004138790.1 | 0.0 | PREDICTED: transcription activator GLK1-like | ||||
| TrEMBL | A0A0A0LQW9 | 0.0 | A0A0A0LQW9_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004158587.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF4242 | 34 | 61 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G20570.1 | 2e-68 | GBF's pro-rich region-interacting factor 1 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.087530.1 |
| Entrez Gene | 101212135 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



