 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.088250.1 | ||||||||
| Common Name | Csa_2G403160 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 373aa MW: 42723.5 Da PI: 7.1521 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 33.4 | 9.8e-11 | 79 | 119 | 3 | 43 |
XXCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 3 elkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNk 43
++k++rr+++NReAAr+sR RKk ++++Le+ +L + +
Cucsa.088250.1 79 DDKVQRRLAQNREAARKSRMRKKVYVQQLETSRLKLAQLEE 119
589***************************85444433333 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 4.2E-8 | 77 | 139 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.335 | 79 | 123 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 3.8E-7 | 80 | 120 | IPR004827 | Basic-leucine zipper domain |
| Gene3D | G3DSA:1.20.5.170 | 3.3E-8 | 81 | 126 | No hit | No description |
| SuperFamily | SSF57959 | 2.89E-7 | 81 | 125 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 84 | 99 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 5.3E-29 | 166 | 241 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0016021 | Cellular Component | integral component of membrane | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 373 aa Download sequence Send to blast |
MIGQQSMSSS STQLYASRMG IYEPFHQINS WPNAFGSRLD TSISPITKVD DCVDNKPEFV 60 PFESMDHLES SQEMNKPIDD KVQRRLAQNR EAARKSRMRK KVYVQQLETS RLKLAQLEEE 120 LERTRQQKGN GCLVDTSHIG FSGLVNPGIA AFEMEYNHWV EEQQRQINEL RKALQVHTTD 180 IELQILVESS LNHYHNLFCM KAKVAKADVF YLMSGVWRSS AERFFLWIGG FRPSELLNVL 240 KPYLEPLNEQ QRADIHKLQQ SSRQAEDALT QGMEKLHQNL SLSIAGDPIG SYISQMGDGM 300 EKFEALESFI SQPSVCYSFL EQADHLRQQT LKRMSHLLTT RQAAQGLLAL GEYFHRLRVL 360 SSLWATRPRE PA* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
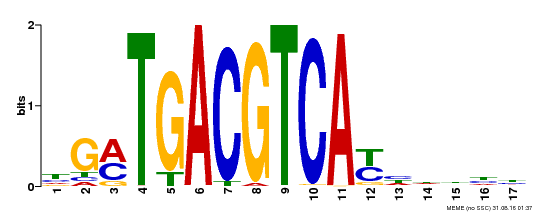
| UniProt | Transcriptional activator that binds specifically to the DNA sequence 5'-TGACG-3'. Recognizes ocs elements like the as-1 motif of the cauliflower mosaic virus 35S promoter. Binding to the as-1-like cis elements mediate auxin- and salicylic acid-inducible transcription. May be involved in the induction of the systemic acquired resistance (SAR) via its interaction with NPR1 (By similarity). {ECO:0000250, ECO:0000269|PubMed:12953119}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00247 | DAP | Transfer from AT1G77920 | Download |
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681815 | 1e-141 | LN681815.1 Cucumis melo genomic scaffold, anchoredscaffold00008. | |||
| GenBank | LN713257 | 1e-141 | LN713257.1 Cucumis melo genomic chromosome, chr_3. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004138645.1 | 0.0 | PREDICTED: transcription factor TGA7-like isoform X1 | ||||
| Refseq | XP_011649910.1 | 0.0 | PREDICTED: transcription factor TGA7-like isoform X1 | ||||
| Refseq | XP_011649911.1 | 0.0 | PREDICTED: transcription factor TGA7-like isoform X1 | ||||
| Swissprot | Q93ZE2 | 1e-139 | TGA7_ARATH; Transcription factor TGA7 | ||||
| TrEMBL | A0A0A0LMH3 | 0.0 | A0A0A0LMH3_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004138645.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF1096 | 34 | 104 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G77920.1 | 1e-137 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.088250.1 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



