 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.102530.1 | ||||||||
| Common Name | Csa_6G491000, LOC101217594 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 416aa MW: 45759.1 Da PI: 5.0435 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 49.4 | 8.1e-16 | 178 | 224 | 4 | 55 |
HHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 4 ahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
hn E+rRR+riN+++ L+ l+P++ K +Ka++L +A+eY+k+Lq
Cucsa.102530.1 178 VHNLSEKRRRSRINEKMKALQNLIPNS-----NKTDKASMLDEAIEYLKQLQ 224
6*************************7.....5******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 3.5E-20 | 171 | 234 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 2.88E-20 | 171 | 234 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 17.86 | 174 | 223 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 7.88E-17 | 177 | 228 | No hit | No description |
| Pfam | PF00010 | 2.7E-13 | 178 | 224 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 8.3E-18 | 180 | 229 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0007623 | Biological Process | circadian rhythm | ||||
| GO:0009409 | Biological Process | response to cold | ||||
| GO:0010114 | Biological Process | response to red light | ||||
| GO:0010154 | Biological Process | fruit development | ||||
| GO:0010187 | Biological Process | negative regulation of seed germination | ||||
| GO:0048440 | Biological Process | carpel development | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 416 aa Download sequence Send to blast |
MDHSVPNNCD RNACSSSTWT LPAPALSSSS SSPPDDISLF LQQILRRSSS SAPHFSLLSP 60 SPSIFSELTC NIRAFTPPTH NLPPPYGPPN AVPDEISAVD SSEQFANSPS SGVLHDPLRT 120 FPTSIPPNAS STSVGASDHN ENDEFDCESE EGLEALVEEL PTKPNPRSSS KRSRAAEVHN 180 LSEKRRRSRI NEKMKALQNL IPNSNKTDKA SMLDEAIEYL KQLQLQVQML SMRNGLSLHP 240 MNLPGSLQYL QLSHMRMDFG EENRSISSDQ ERPNQIFLSL PDQKAASIHP FMSDIGRTNA 300 AETPFELVPP IQAHLVPFYL SESSKSKEIC SRDLLRDDHQ AVNVNVSQSE TTPLVSCPYN 360 IPETPDLQGL QNGLSVEPCI IEGNRSGVLL NYTPEHSLVF PSPFNGIKQE DRLKQ* |
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00035 | PBM | Transfer from AT4G36930 | Download |
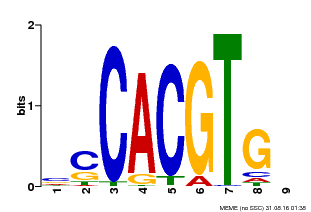
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681875 | 0.0 | LN681875.1 Cucumis melo genomic scaffold, anchoredscaffold00007. | |||
| GenBank | LN713262 | 0.0 | LN713262.1 Cucumis melo genomic chromosome, chr_8. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004142005.2 | 0.0 | PREDICTED: transcription factor SPATULA | ||||
| TrEMBL | A0A0A0KLD3 | 0.0 | A0A0A0KLD3_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004157205.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF2276 | 33 | 79 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT4G36930.1 | 2e-39 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.102530.1 |
| Entrez Gene | 101217594 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



