 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.149820.1 | ||||||||
| Common Name | Csa_3G637990, LOC101219098 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | G2-like | ||||||||
| Protein Properties | Length: 262aa MW: 29539.6 Da PI: 6.1636 | ||||||||
| Description | G2-like family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | G2-like | 104.3 | 7.4e-33 | 18 | 72 | 1 | 55 |
G2-like 1 kprlrWtpeLHerFveaveqLGGsekAtPktilelmkvkgLtlehvkSHLQkYRl 55
kprlrWt +LH+rFv+av++LGG+ekAtPk++l+lm++kgLtl+h+kSHLQkYRl
Cucsa.149820.1 18 KPRLRWTADLHDRFVDAVTKLGGPEKATPKSVLRLMGLKGLTLYHLKSHLQKYRL 72
79****************************************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.60 | 7.8E-32 | 15 | 75 | IPR009057 | Homeodomain-like |
| PROSITE profile | PS51294 | 11.609 | 15 | 75 | IPR017930 | Myb domain |
| SuperFamily | SSF46689 | 5.55E-16 | 18 | 75 | IPR009057 | Homeodomain-like |
| TIGRFAMs | TIGR01557 | 3.9E-24 | 18 | 72 | IPR006447 | Myb domain, plants |
| Pfam | PF00249 | 7.5E-9 | 20 | 71 | IPR001005 | SANT/Myb domain |
| Pfam | PF14379 | 3.0E-19 | 105 | 152 | IPR025756 | MYB-CC type transcription factor, LHEQLE-containing domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 262 aa Download sequence Send to blast |
MERNYPYENG VMMTRDPKPR LRWTADLHDR FVDAVTKLGG PEKATPKSVL RLMGLKGLTL 60 YHLKSHLQKY RLGLQTRKQN VAEQRNESSG TLSNFSGVEE DDRGMQIAEA LKSHVEVQKT 120 ILEQLEVQNK LQMRIEAQGK YLQDILENAQ KSLALAINSN LGSLDQSTEM QLINFDAALS 180 DQIEKFKKQE IRGSITNLND VCKKTGDSLI QICKMEAAEE DTNEFNIEKN MINFDLNSKG 240 GYDYSANGAE ILEPKVLPCS R* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 6j4k_A | 7e-20 | 18 | 71 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4k_B | 7e-20 | 18 | 71 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_A | 7e-20 | 18 | 71 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_B | 7e-20 | 18 | 71 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_C | 7e-20 | 18 | 71 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j4r_D | 7e-20 | 18 | 71 | 1 | 54 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_A | 7e-20 | 18 | 71 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_C | 7e-20 | 18 | 71 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_D | 7e-20 | 18 | 71 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_F | 7e-20 | 18 | 71 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_H | 7e-20 | 18 | 71 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| 6j5b_J | 7e-20 | 18 | 71 | 2 | 55 | Protein PHOSPHATE STARVATION RESPONSE 1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00544 | DAP | Transfer from AT5G45580 | Download |
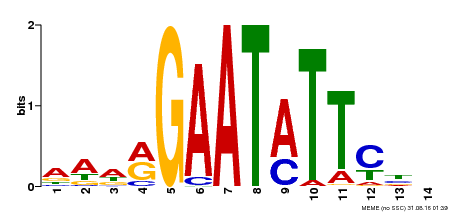
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681829 | 0.0 | LN681829.1 Cucumis melo genomic scaffold, anchoredscaffold00054. | |||
| GenBank | LN713258 | 0.0 | LN713258.1 Cucumis melo genomic chromosome, chr_4. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004144408.1 | 0.0 | PREDICTED: myb family transcription factor APL | ||||
| Swissprot | C0SVS4 | 1e-69 | PHLB_ARATH; Myb family transcription factor PHL11 | ||||
| TrEMBL | A0A0A0LCI1 | 0.0 | A0A0A0LCI1_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004160999.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF5101 | 31 | 42 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G45580.1 | 3e-70 | G2-like family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.149820.1 |
| Entrez Gene | 101219098 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



