 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.159820.1 | ||||||||
| Common Name | Csa_2G367240, LOC101211469 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | E2F/DP | ||||||||
| Protein Properties | Length: 307aa MW: 35438.9 Da PI: 7.5007 | ||||||||
| Description | E2F/DP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | E2F_TDP | 81.9 | 5.5e-26 | 50 | 115 | 1 | 71 |
E2F_TDP 1 rkeksLrlltqkflkllekseegivtlnevakeLvsedvknkrRRiYDilNVLealnliekkekneirwkg 71
r+++sL+ lt+kf++l++++e+g+++ln+ a+ L +v ++RRiYDi+NVLe+++liek+ n+irwkg
Cucsa.159820.1 50 RYDSSLGFLTKKFIRLVQEAEDGTLDLNKTADVL---KV--QKRRIYDITNVLEGIGLIEKTTTNHIRWKG 115
6899******************************...88..****************************98 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:1.10.10.10 | 3.5E-27 | 46 | 116 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SuperFamily | SSF46785 | 5.85E-17 | 50 | 113 | IPR011991 | Winged helix-turn-helix DNA-binding domain |
| SMART | SM01372 | 1.6E-33 | 50 | 115 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| Pfam | PF02319 | 1.4E-24 | 52 | 115 | IPR003316 | E2F/DP family, winged-helix DNA-binding domain |
| CDD | cd14660 | 5.38E-36 | 126 | 230 | No hit | No description |
| SuperFamily | SSF144074 | 6.8E-27 | 127 | 230 | No hit | No description |
| Pfam | PF16421 | 2.0E-25 | 131 | 230 | IPR032198 | E2F transcription factor, CC-MB domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0000902 | Biological Process | cell morphogenesis | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0042023 | Biological Process | DNA endoreduplication | ||||
| GO:0051782 | Biological Process | negative regulation of cell division | ||||
| GO:0005667 | Cellular Component | transcription factor complex | ||||
| GO:0005730 | Cellular Component | nucleolus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| GO:0046982 | Molecular Function | protein heterodimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 307 aa Download sequence Send to blast |
MRSNDSSCEP VSSANEKQNK KFKLQKNSKS KTRKSIDEPV DSPNQSTNGR YDSSLGFLTK 60 KFIRLVQEAE DGTLDLNKTA DVLKVQKRRI YDITNVLEGI GLIEKTTTNH IRWKGGERRG 120 PQELNDQVGR LKDEVKSLYA DERRLDELIR MKQELLRNLE QNAHYRNHLF ITEEDILRIP 180 CFKNQTLIAV KAPQASCIEV PDPDEEACFS ERQCRMIIKS TTGPIDLYLL RTAKQELEEN 240 TSKQAKLCLA QQKNPNIFTN NTYSPFQDLH GMQRILPLHN NIDDDYWFQS NSQVSITHLW 300 GEEHNF* |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 1cf7_A | 2e-23 | 46 | 115 | 3 | 73 | PROTEIN (TRANSCRIPTION FACTOR E2F-4) |
| Search in ModeBase | ||||||
| Nucleic Localization Signal ? help Back to Top | |||
|---|---|---|---|
| No. | Start | End | Sequence |
| 1 | 82 | 88 | LKVQKRR |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Involved in transcriptional repression. May act by repressing E2F-regulated genes in mature differentiated cells, but is not an antagonist of E2FA. Restricts cell division and is involved in the coordination between cell proliferation and endoreduplication during development. May play a role during the transition from skotomorphogenesis to photomorphogenesis. Regulated by phosphorylation-dependent proteolysis via the protein-ubiquitin ligase SCF(SKP2A) complex. {ECO:0000269|PubMed:11786543, ECO:0000269|PubMed:11891240, ECO:0000269|PubMed:12468727, ECO:0000269|PubMed:16920782, ECO:0000269|PubMed:19662336}. | |||||
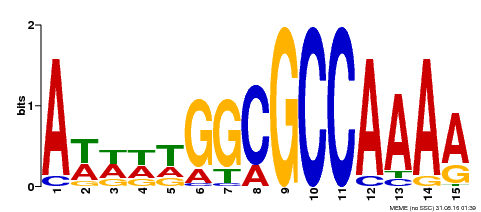
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00190 | DAP | Transfer from AT1G47870 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Down-regulated by light. {ECO:0000269|PubMed:12468727, ECO:0000269|PubMed:18424613}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681823 | 5e-60 | LN681823.1 Cucumis melo genomic scaffold, anchoredscaffold00014. | |||
| GenBank | LN713257 | 5e-60 | LN713257.1 Cucumis melo genomic chromosome, chr_3. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004152759.1 | 0.0 | PREDICTED: transcription factor E2FC isoform X1 | ||||
| Swissprot | Q9FV70 | 1e-86 | E2FC_ARATH; Transcription factor E2FC | ||||
| TrEMBL | A0A0A0LL68 | 0.0 | A0A0A0LL68_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004152759.1 | 0.0 | (Cucumis sativus) | ||||
| STRING | XP_004167900.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF11885 | 25 | 29 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G47870.2 | 1e-88 | E2F/DP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.159820.1 |
| Entrez Gene | 101211469 |



