 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.175390.1 | ||||||||
| Common Name | Csa_3G734310, LOC101207082 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | Dof | ||||||||
| Protein Properties | Length: 345aa MW: 36502.5 Da PI: 9.8407 | ||||||||
| Description | Dof family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | zf-Dof | 123.9 | 5.2e-39 | 79 | 138 | 3 | 62 |
zf-Dof 3 ekalkcprCdstntkfCyynnyslsqPryfCkaCrryWtkGGalrnvPvGggrrknkkss 62
e+alkcprC+stntkfCy+nnysl+qPr+fCk+CrryWt+GGalrnvPvGgg+r+nk+s+
Cucsa.175390.1 79 EAALKCPRCESTNTKFCYFNNYSLTQPRHFCKTCRRYWTRGGALRNVPVGGGCRRNKRSK 138
7789*****************************************************975 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SuperFamily | SSF101447 | 3.92E-5 | 39 | 49 | No hit | No description |
| ProDom | PD007478 | 3.0E-31 | 62 | 127 | IPR003851 | Zinc finger, Dof-type |
| Pfam | PF02701 | 1.3E-32 | 81 | 136 | IPR003851 | Zinc finger, Dof-type |
| PROSITE profile | PS50884 | 29.544 | 82 | 136 | IPR003851 | Zinc finger, Dof-type |
| PROSITE pattern | PS01361 | 0 | 84 | 120 | IPR003851 | Zinc finger, Dof-type |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 345 aa Download sequence Send to blast |
MNFSSIPPYL DPSNWQQQVT HQVGTSSSTA LSSQLLPPPP PPPPPPPPPL PPHGVGGAGS 60 IRPGSMAERA RMANIPMPEA ALKCPRCEST NTKFCYFNNY SLTQPRHFCK TCRRYWTRGG 120 ALRNVPVGGG CRRNKRSKGS SSKSPPVSSD RQQTSGSANS SSSAIASNNS GGLSPQIPPL 180 GRFMAPLHQQ FSDFDIGSYS YGGGLSAPAS ATGDLSFQLG NTNLGGGTSI GSLLGFDHQQ 240 WRLQQQPPQF PFLSSLDPFE GGNGGGGEAP GTAWQMRPKA PSTSSRNNLT QMGNSVKMEE 300 TPDQVNNNVG RQSIGINEQY WSSGSMAWSD LSGFSSSSST RNPL* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Transcription factor that binds specifically to a 5'-AA[AG]G-3' consensus core sequence (By similarity). Probably involved in early processes for vascular development (PubMed:17583520). The PEAR proteins (e.g. DOF2.4, DOF5.1, DOF3.2, DOF1.1, DOF5.6 and DOF5.3) activate gene expression that promotes radial growth of protophloem sieve elements. Triggers the transcription of HD-ZIP III genes, especially in the central domain of vascular tissue (PubMed:30626969). {ECO:0000250|UniProtKB:Q9M2U1, ECO:0000269|PubMed:17583520, ECO:0000269|PubMed:30626969}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00295 | DAP | Transfer from AT2G37590 | Download |
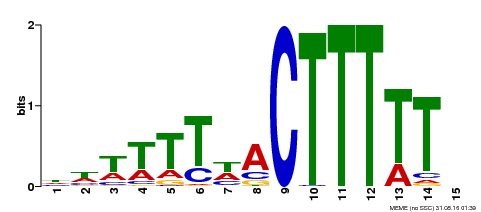
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: By cytokinin in procambium. Antagonized by the HD-ZIP III proteins and by mobile miR165 and miR166 microRNAs. {ECO:0000269|PubMed:30626969}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681841 | 0.0 | LN681841.1 Cucumis melo genomic scaffold, anchoredscaffold00011. | |||
| GenBank | LN713258 | 0.0 | LN713258.1 Cucumis melo genomic chromosome, chr_4. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004137942.1 | 0.0 | PREDICTED: dof zinc finger protein DOF5.1-like | ||||
| Swissprot | O80928 | 5e-52 | DOF24_ARATH; Dof zinc finger protein DOF2.4 | ||||
| TrEMBL | A0A0A0LAK9 | 0.0 | A0A0A0LAK9_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004137942.1 | 0.0 | (Cucumis sativus) | ||||
| STRING | XP_004173251.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF656 | 34 | 143 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT2G37590.1 | 1e-42 | DNA binding with one finger 2.4 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.175390.1 |
| Entrez Gene | 101207082 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



