 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.198520.1 | ||||||||
| Common Name | Csa_1G015080, LOC101211973 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | bHLH | ||||||||
| Protein Properties | Length: 560aa MW: 61190 Da PI: 8.8034 | ||||||||
| Description | bHLH family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | HLH | 53.6 | 4e-17 | 259 | 308 | 2 | 55 |
HHHHHHHHHHHHHHHHHHHHHHHCTSCCC...TTS-STCHHHHHHHHHHHHHHH CS
HLH 2 rrahnerErrRRdriNsafeeLrellPkaskapskKlsKaeiLekAveYIksLq 55
r +h++ E+rRR++iN++f+ Lr l+P++ ++K +Ka+ L ++veYI+ Lq
Cucsa.198520.1 259 RSKHSATEQRRRSKINDRFQMLRGLIPHS----DQKRDKASFLLEVVEYIQFLQ 308
889*************************9....9*******************9 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:4.10.280.10 | 3.2E-25 | 254 | 329 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SuperFamily | SSF47459 | 5.89E-19 | 256 | 327 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| PROSITE profile | PS50888 | 16.437 | 257 | 307 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| CDD | cd00083 | 8.06E-15 | 259 | 312 | No hit | No description |
| Pfam | PF00010 | 1.3E-14 | 259 | 308 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| SMART | SM00353 | 3.2E-13 | 263 | 313 | IPR011598 | Myc-type, basic helix-loop-helix (bHLH) domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006351 | Biological Process | transcription, DNA-templated | ||||
| GO:0009742 | Biological Process | brassinosteroid mediated signaling pathway | ||||
| GO:1902448 | Biological Process | positive regulation of shade avoidance | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003677 | Molecular Function | DNA binding | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0046983 | Molecular Function | protein dimerization activity | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 560 aa Download sequence Send to blast |
MELPQPRPFG AEGSKSTHDF LSLYTHSSPQ LDPRSTPQGS YLKTHDFLQP QERIRKASTK 60 EETDVERPPP PAPPPSVEHI LPGGIGTYSI SHVSYFDQRV VLPKPEGSVF TGVRSSSSAE 120 RNDENSNCSS FAAAGSGFTL WEESSVKKGK TGKENNVGDR PHEPRASTSQ WTASMERPSQ 180 SSSNNHHNTF SCLSSSQPTG TKNPTFMEML KSAKSTSQDE ELDDDGDFVI KKETSTANKG 240 GLRIKVDGNS SDQKANTPRS KHSATEQRRR SKINDRFQML RGLIPHSDQK RDKASFLLEV 300 VEYIQFLQEK VQKYEGSYQE WNHEMAKLVP LRNNQRSADV YNDQSRGINS GSVPALVLAA 360 KFIEKNSPLS PIVPGSAHNA VDSDTSSAST LKAVDHHSGR TSNAVQFPMS IPPKLSASTR 420 DGNVVPQPPK PLSSGMDHSS LRPEIRSCEA RCFNSDVAVA SEMQKEQDLT IEGGTINISS 480 VYSQGLLNTL THALQSSGVD LSQARISVQI ELGKRASRRA ISPASIVKDA NDMGMMHARV 540 SGTEDSERAT KKLKTTMKN* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Positive brassinosteroid-signaling protein. Transcription factor that bind specifically to the DNA sequence 5'-CANNTG-3'(E box). Can bind individually to the promoter as a homodimer or synergistically as a heterodimer with BZR2/BES1. Does not itself activate transcription but enhances BZR2/BES1-mediated target gene activation. {ECO:0000269|PubMed:15680330}. | |||||
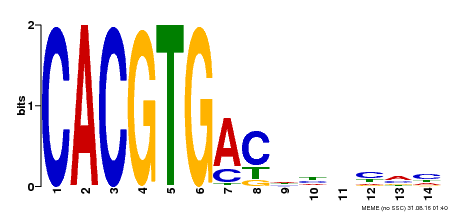
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00498 | DAP | Transfer from AT5G08130 | Download |
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Repressed by cold treatment. {ECO:0000269|PubMed:12679534}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681813 | 0.0 | LN681813.1 Cucumis melo genomic scaffold, anchoredscaffold00030. | |||
| GenBank | LN713256 | 0.0 | LN713256.1 Cucumis melo genomic chromosome, chr_2. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011648566.1 | 0.0 | PREDICTED: transcription factor BIM1 | ||||
| Swissprot | Q9LEZ3 | 1e-113 | BIM1_ARATH; Transcription factor BIM1 | ||||
| TrEMBL | A0A0A0LSI8 | 0.0 | A0A0A0LSI8_CUCSA; Uncharacterized protein | ||||
| STRING | XP_004138212.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF7252 | 31 | 45 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G08130.4 | 1e-117 | bHLH family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.198520.1 |
| Entrez Gene | 101211973 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



