 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.199800.3 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | bZIP | ||||||||
| Protein Properties | Length: 379aa MW: 42738.4 Da PI: 7.4834 | ||||||||
| Description | bZIP family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | bZIP_1 | 35 | 3.1e-11 | 72 | 113 | 4 | 45 |
XCHHHCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHH CS
bZIP_1 4 lkrerrkqkNReAArrsRqRKkaeieeLeekvkeLeaeNkaL 45
k rr+++NReAAr+sR+RKka+i++Le +L++ ++L
Cucsa.199800.3 72 AKTMRRLAQNREAARKSRLRKKAYIQQLESSRIKLSQLEQDL 113
6889*************************9777777666655 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| SMART | SM00338 | 9.5E-7 | 69 | 135 | IPR004827 | Basic-leucine zipper domain |
| PROSITE profile | PS50217 | 9.128 | 71 | 115 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF00170 | 2.2E-8 | 72 | 113 | IPR004827 | Basic-leucine zipper domain |
| SuperFamily | SSF57959 | 3.38E-7 | 73 | 114 | No hit | No description |
| CDD | cd14708 | 7.88E-21 | 73 | 124 | No hit | No description |
| Gene3D | G3DSA:1.20.5.170 | 4.1E-8 | 74 | 116 | No hit | No description |
| PROSITE pattern | PS00036 | 0 | 76 | 91 | IPR004827 | Basic-leucine zipper domain |
| Pfam | PF14144 | 1.6E-31 | 156 | 230 | IPR025422 | Transcription factor TGA like domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0006355 | Biological Process | regulation of transcription, DNA-templated | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 379 aa Download sequence Send to blast |
MESESPISRR TCSSNQGLFD QNHHHHLLHL QHLQSEFEDD ALRTEPSSQQ NQSPPKEKRK 60 GGGSTSERQL DAKTMRRLAQ NREAARKSRL RKKAYIQQLE SSRIKLSQLE QDLHRARSQG 120 LFLGACGGVM GGNISSGAAI FDMEYARWLD EDHRLMAELR AALQGHLPDG DLRAIVDSYI 180 SHYDEIFHLK GVAAKSDVFH LITGMWMTPA ERCFLWIGGF RPSKLIEMLV PQIDTLTDQQ 240 ALGICNLQRS SQETEDALYQ GLEQLQHSLI ITIAGTAVVD GINHMALAAG KLSNLEGFIR 300 QADMLRQQTL HQLHRILTVR QAARCFVVIG EYYGRLRALS SLWVSRPRES SCLNDESSCQ 360 TTTELQMIQN SHTHFPNF* |
| Functional Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | Together with TGA10, basic leucine-zipper transcription factor required for anther development, probably via the activation of SPL expression in anthers and via the regulation of genes with functions in early and middle tapetal development (PubMed:20805327). Required for signaling responses to pathogen-associated molecular patterns (PAMPs) such as flg22 that involves chloroplastic reactive oxygen species (ROS) production and subsequent expression of H(2)O(2)-responsive genes (PubMed:27717447). {ECO:0000269|PubMed:20805327, ECO:0000269|PubMed:27717447}. | |||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00131 | DAP | Transfer from AT1G08320 | Download |
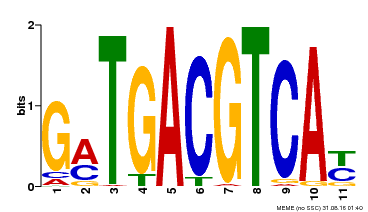
| |||
| Regulation -- Description ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Description | |||||
| UniProt | INDUCTION: Induced by flg22 in leaves. {ECO:0000269|PubMed:27717447}. | |||||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681928 | 1e-122 | LN681928.1 Cucumis melo genomic scaffold, anchoredscaffold00004. | |||
| GenBank | LN713266 | 1e-122 | LN713266.1 Cucumis melo genomic chromosome, chr_12. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_011654412.1 | 0.0 | PREDICTED: transcription factor HBP-1b(c38) | ||||
| Swissprot | Q93XM6 | 1e-172 | TGA9_ARATH; Transcription factor TGA9 | ||||
| TrEMBL | A0A1S3CEU5 | 0.0 | A0A1S3CEU5_CUCME; transcription factor HBP-1b(C38)-like | ||||
| STRING | XP_004139481.1 | 0.0 | (Cucumis sativus) | ||||
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT1G08320.2 | 1e-175 | bZIP family protein | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.199800.3 |
| Publications ? help Back to Top | |||
|---|---|---|---|
|
|||



