 |
PlantRegMap/PlantTFDB v5.0
Plant Transcription
Factor Database
|
| Home TFext BLAST Prediction Download Help About Links PlantRegMap |
Transcription Factor Information
| Basic Information? help Back to Top | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| TF ID | Cucsa.201710.1 | ||||||||
| Organism | |||||||||
| Taxonomic ID | |||||||||
| Taxonomic Lineage |
cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliophyta; Mesangiospermae; eudicotyledons; Gunneridae; Pentapetalae; rosids; fabids; Cucurbitales; Cucurbitaceae; Benincaseae; Cucumis
|
||||||||
| Family | WRKY | ||||||||
| Protein Properties | Length: 326aa MW: 35890.5 Da PI: 6.3393 | ||||||||
| Description | WRKY family protein | ||||||||
| Gene Model |
|
||||||||
| Signature Domain? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| No. | Domain | Score | E-value | Start | End | HMM Start | HMM End |
| 1 | WRKY | 96.5 | 1.8e-30 | 175 | 233 | 2 | 59 |
--SS-EEEEEEE--TT-SS-EEEEEE-ST.T---EEEEEE-SSSTTEEEEEEES--SS- CS
WRKY 2 dDgynWrKYGqKevkgsefprsYYrCtsa.gCpvkkkversaedpkvveitYegeHnhe 59
D ++WrKYGqK++kgs++pr+YYrC+s+ gC ++k+vers++dp++++itY+g+H h+
Cucsa.201710.1 175 TDMWAWRKYGQKPIKGSPYPRNYYRCSSSkGCGARKQVERSNDDPETFTITYTGDHSHP 233
69*************************998****************************8 PP
| |||||||
| Protein Features ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Database | Entry ID | E-value | Start | End | InterPro ID | Description |
| Gene3D | G3DSA:2.20.25.80 | 4.3E-30 | 160 | 235 | IPR003657 | WRKY domain |
| PROSITE profile | PS50811 | 32.214 | 169 | 235 | IPR003657 | WRKY domain |
| SuperFamily | SSF118290 | 4.45E-26 | 169 | 235 | IPR003657 | WRKY domain |
| SMART | SM00774 | 3.0E-36 | 174 | 234 | IPR003657 | WRKY domain |
| Pfam | PF03106 | 3.8E-26 | 176 | 233 | IPR003657 | WRKY domain |
| Gene Ontology ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| GO Term | GO Category | GO Description | ||||
| GO:0007263 | Biological Process | nitric oxide mediated signal transduction | ||||
| GO:0009739 | Biological Process | response to gibberellin | ||||
| GO:0042742 | Biological Process | defense response to bacterium | ||||
| GO:0045892 | Biological Process | negative regulation of transcription, DNA-templated | ||||
| GO:0005634 | Cellular Component | nucleus | ||||
| GO:0003700 | Molecular Function | transcription factor activity, sequence-specific DNA binding | ||||
| GO:0043565 | Molecular Function | sequence-specific DNA binding | ||||
| GO:0044212 | Molecular Function | transcription regulatory region DNA binding | ||||
| Sequence ? help Back to Top |
|---|
| Protein Sequence Length: 326 aa Download sequence Send to blast |
MAAGNDDWDL SAVVRSCNSA GSATDPTSAA AEESALSCLA SLTFDDDPND VAFSFSDIFQ 60 PKQPNGFHEL HQAFISFLPN PSTTATTVTT VPAAEPEIPY LTTPPTNRHF RQVMKPIRPN 120 PHPHPVALHH HHRQPPFSPD LPNSPMTHSL IPKSRKRKNQ QKRRVCHVTA DNLSTDMWAW 180 RKYGQKPIKG SPYPRNYYRC SSSKGCGARK QVERSNDDPE TFTITYTGDH SHPRPTHRNS 240 LAGSSRNRSS SSSSSSRHPT MGDSDPPMMT ASVLLPSSSS PAASPITPLN DYDSTIGEKD 300 GEMFEDMPID SDDEDDDDDI LIPNLT |
| 3D Structure ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| PDB ID | Evalue | Query Start | Query End | Hit Start | Hit End | Description |
| 5w3x_B | 6e-19 | 176 | 236 | 18 | 80 | Disease resistance protein RRS1 |
| 5w3x_D | 6e-19 | 176 | 236 | 18 | 80 | Disease resistance protein RRS1 |
| Search in ModeBase | ||||||
| Binding Motif ? help Back to Top | |||
|---|---|---|---|
| Motif ID | Method | Source | Motif file |
| MP00561 | DAP | Transfer from AT5G52830 | Download |
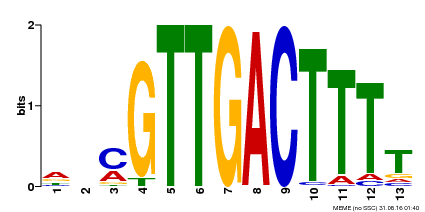
| |||
| Regulation -- PlantRegMap ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Upstream Regulator | Target Gene | ||||
| PlantRegMap | Retrieve | Retrieve | ||||
| Annotation -- Nucleotide ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | |||
| GenBank | LN681895 | 1e-136 | LN681895.1 Cucumis melo genomic scaffold, anchoredscaffold00005. | |||
| GenBank | LN713263 | 1e-136 | LN713263.1 Cucumis melo genomic chromosome, chr_9. | |||
| Annotation -- Protein ? help Back to Top | |||||||
|---|---|---|---|---|---|---|---|
| Source | Hit ID | E-value | Description | ||||
| Refseq | XP_004145903.1 | 0.0 | PREDICTED: probable WRKY transcription factor 27 | ||||
| TrEMBL | A0A1S3AU96 | 1e-171 | A0A1S3AU96_CUCME; probable WRKY transcription factor 27 isoform X1 | ||||
| STRING | XP_004145903.1 | 0.0 | (Cucumis sativus) | ||||
| STRING | XP_004160353.1 | 0.0 | (Cucumis sativus) | ||||
| Orthologous Group ? help Back to Top | |||
|---|---|---|---|
| Lineage | Orthologous Group ID | Taxa Number | Gene Number |
| Fabids | OGEF6794 | 32 | 49 |
| Best hit in Arabidopsis thaliana ? help Back to Top | ||||||
|---|---|---|---|---|---|---|
| Hit ID | E-value | Description | ||||
| AT5G52830.1 | 3e-45 | WRKY DNA-binding protein 27 | ||||
| Link Out ? help Back to Top | |
|---|---|
| Phytozome | Cucsa.201710.1 |



